The field of protein engineering and drug discovery now benefits from mRNA display technology as an effective means to identify proteins and antibodies with high affinity alongside peptides. Custom mRNA display libraries represent the fundamental element of this technology because they enable the identification of functional molecules. This blog post examines the techniques and innovative approaches used in developing mRNA display libraries alongside important considerations while explaining how these libraries propel biotechnology and therapeutic advancements.
Introduction to mRNA Display Technology
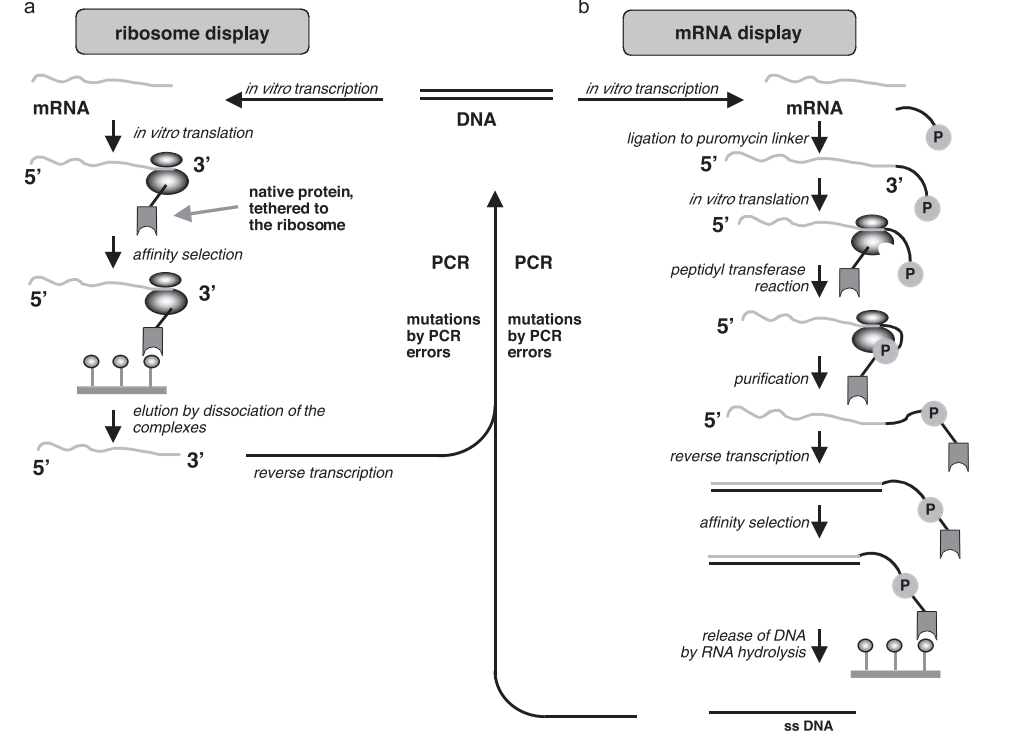
mRNA display serves as a sturdy in vitro selection tool which creates a stable connection between genotype (mRNA) and phenotype (encoded protein) through mRNA-protein fusion. The stable mRNA-protein fusion enables scientists to test extensive protein and peptide libraries for their binding abilities and functional characteristics. The mRNA display technique functions outside living cells which allows researchers to identify molecules that traditional in vivo methods cannot because those molecules would be toxic or hard to express in living organisms.
mRNA display enables the exploration of massive sequence libraries because they can hold up to 10¹⁵ unique sequences which surpasses the diversity available with conventional techniques. The most significant advantage of mRNA display originates from its ability to create custom libraries which enable researchers to target precise scientific or medical objectives. Successful optimization of antibody binding and enzyme evolution as well as the discovery of new ligands require strategic mRNA display library construction to realize their full capabilities. For more in – depth understanding of mRNA display screening, you can visit mRNA Display Screening Services.

Selection and evolution of proteins by mRNA display(Blanco, et al et al. 2020)
Core Strategies for Constructing mRNA Display Libraries
Developing a functional mRNA display library demands meticulous attention to its diversity and complex structure alongside its practicality. The following section outlines the fundamental strategies used for library design.
2.1 Barcoded Antigen Libraries: Enhancing Tracking and Diversity
Antigen libraries with barcodes consist of unique nucleotide tags embedded within mRNA sequences to allow multiplexed screening and exact molecular tracking. Researchers achieve simultaneous molecular screening of thousands of antigens or peptide variants and post-selection origin tracing by attaching barcodes to each variant.
- How Barcoding Works:Barcodes represent short synthetic DNA fragments that researchers integrate into mRNA constructs. These molecular identifiers serve as barcodes that permit high-throughput sequencing to measure variant abundance before and after selection.
Advantages:
- Multiplexing:Parallel screening of multiple libraries or conditions maximizes efficiency by reducing both time and resource usage.
- Quantitative Analysis: Barcodes deliver precise tracking of sequence enrichment and depletion which enhances the clarity of selection results.
- Complexity Boost:Libraries maintain tractability while increasing effective diversity through the combination of barcodes and multiple antigen sequences.
2.2 Mutagenic PCR: Expanding Sequence Space
Through the introduction of random mutations, mutagenic PCR generates numerous variants of target genes or peptide-encoding sequences. This technique enables scientists to identify amino acid changes that enhance binding strength and stability along with additional functional traits.
- Optimizing Mutation Rates: The goal is to balance mutation rates so they prevent excessive non-functional sequences while preserving sufficient diversity for successful results. Researchers can adjust PCR mutation rates by using error-prone PCR enzymes or changing reaction conditions with additions like manganese ions.
- Applications:
- Through directed evolution scientists work to improve the catalytic efficiency of enzymes, as demonstrated in mRNA Display-based Enzyme Directed Evolution..
- Affinity maturation of antibodies requires mutations in the complementarity-determining regions (CDRs), which is also related to mRNA Display-based Antibody Affinity Maturation..
- Study peptide libraries to identify novel ligands and inhibitor molecules, similar to the services provided in mRNA Display-based Peptide Discovery..
2.3 Designing Peptide and Nanobody Libraries
Unlike general mRNA display libraries, peptide and nanobody libraries represent specialized subsets engineered for particular applications.
- Peptide Library Design:
- Length and Motifs: Peptide sequences between 6 and 30 amino acids exhibit high binding specificity when they are shorter and enable complex structural formations when they extend to longer lengths.
- Diversity:Researchers create libraries by fully randomizing sequences or partially randomizing them to maintain functional motifs or by biasing amino acid distributions to replicate natural binding domains. For peptide library – related services, mRNA Display-based Peptide Library Construction offers comprehensive solutions.
- Nanobody Library Construction:
- CDR Diversification: Nanobodies (single-domain antibodies from camelids) rely on hypervariable complementarity-determining regions (CDRs) for antigen binding. Libraries are often constructed by diversifying CDR3, the most variable region.
- Stability Optimization: Incorporating framework region mutations can improve nanobody solubility and stability without compromising binding affinity.
Table: Comparison of Key mRNA Display Library Construction Strategies
| Strategy | Key Features | Advantages | Applications |
| Barcoded Antigen Libraries | Integration of unique nucleotide tags for multiplexed tracking. | Enables high-throughput screening, quantitative analysis, and enhanced diversity. | Antibody discovery, ligand profiling. |
| Mutagenic PCR | Random mutagenesis to expand sequence space. | Generates diverse variants for directed evolution and affinity maturation. | Enzyme engineering, antibody optimization. |
| Peptide Libraries | Short, randomized peptide sequences (6–30 amino acids). | High binding specificity; suitable for ligand discovery and inhibitor design. | Drug targeting, biomarker recognition. |
| Nanobody Libraries | Diversification of CDR regions in single-domain antibodies. | Small size, high stability, and ability to target cryptic epitopes. | Therapeutic antibody development, diagnostics. |
The table provides insights into each strategy’s strengths and applications to assist researchers in choosing suitable methods for their objectives.
Scientists who adopt these strategies and innovations will be able to maximize the capabilities of mRNA display to create significant advancements in biotechnology and medical research.
Optimizing Library Size and Quality
The effectiveness of screening in mRNA display libraries depends on both their size and quality. The potential diversity of larger libraries must be balanced with practical constraints such as synthesis expenses and screening capacity.
3.1 Library Size Considerations
- Theoretical vs. Practical Limits:Most applications function best with a library size ranging from 10¹²–10¹⁵ because this range includes most peptide or antibody sequences. Building libraries larger than 10¹⁴ demands advanced synthesis techniques.
- Functional Representation: An effective protein library should enable the translation of diverse sequences into functional proteins with high efficiency. Effective diversity suffers when DNA synthesis or mRNA translation introduces biases which makes careful validation a crucial step.
3.2 Validating Library Complexity
- Next-Generation Sequencing (NGS):Researchers can learn about library diversity and mutation patterns along with potential biases by sequencing part of the library before selection.
- Clonal Analysis:Sequencing individual clones reveals sequences that appear more or less frequently than expected.
- Functional Assays: Functionally relevant diversity in a library can be verified by testing a small subset for binding or activity.
3.3 Balancing Diversity and Functionality
Excessive diversification often results in large numbers of proteins that fail to fold correctly or lack functionality. Strategies to mitigate this include:
- Semi-randomization: The semi-randomization approach restricts randomization to certain areas like CDRs in antibodies but maintains essential structural domains.
- Stability Filters: To promote proper protein folding during library construction scientists use chaperones and solubility tags as supportive elements.
Innovations in mRNA Display Library Design
Synthetic biology advancements coupled with computational tools are transforming both the design and implementation of mRNA display libraries.
4.1 Combinatorial Tagging and Iterative Diversification
Combinatorial tagging requires dividing a library into different subpools where each subpool receives a distinct barcode label. This method enables researchers to examine sequence space with reduced complexity in each selection cycle through iterative diversification and screening of subpools.
4.2 Computational Design Integration
Researchers now frequently employ machine learning algorithms and molecular modeling as computational tools to identify top candidate sequences ahead of library creation. For example:
- Predicting peptide binding motifs using structural data.
- Finding nanobody structures which effectively stabilize various CDR3 regions.
- Optimizing library diversity by simulating sequence distributions.
4.3 Applications in Emerging Fields
- Antibody Discovery:Researchers employ custom libraries to identify antibodies that target complex structures like membrane proteins and fast-changing viruses, as described inmRNA Display-based Antibody Discovery..
- Enzyme Evolution:Mutagenic PCR and site-directed mutagenesis techniques used in library design allow for enzyme evolution that produces both enhanced activity and new functions, similar to the services provided inmRNA Display-based Enzyme Directed Evolution..
- Diagnostics:Peptide libraries enable the identification of ligands that attach to disease biomarkers which aids in creating new diagnostic assays, related tomRNA Display-based Peptide Discovery..
Challenges and Future Directions
The construction process of mRNA display libraries includes multiple obstacles despite its successful applications.
- Aggregation and Instability: During in vitro translation some proteins and peptides develop aggregates that lead to lower quality library outcomes.
- Transfection Efficiency:The ability to convert mRNA into proteins in cell-free systems presents a major obstacle for scaling up large libraries.
- Cost and Scalability:The expense of building and examining libraries that exceed 10¹⁴ members becomes economically impractical.
The advancement of synthetic biology along with automation technology holds promise for solving the existing challenges.
- Improved Cell-Free Systems: Translation platforms now offer enhanced performance by providing better stability alongside improved efficiency.
- High-Throughput Screening:Developments in microfluidics and robotics will lead to quicker and less expensive screening of extensive libraries.
- AI-Driven Design: Through machine learning models we can optimize library design by predicting the best sequences which will minimize trial-and-error experimentation.
Conclusion
The cutting-edge technology of custom mRNA display libraries leads protein engineering forward by providing unmatched possibilities to identify and enhance molecules for scientific research and therapeutic applications. Researchers can build targeted libraries by applying techniques like barcoding, mutagenic PCR, and computational design to maintain a balance between diversity, functionality, and practicality. Advancing technology reveals promising prospects for mRNA display libraries which can revolutionize both drug discovery processes and enzyme development. CD Biosynsis offers a wide range of services related to mRNA display libraries, including antibody discovery, protein discovery, and peptide – related services. You can explore these services to find the most suitable solutions for your research needs.
Reference
Blanco, Celia, et al. “High throughput sequencing of in vitro selections of mRNA-displayed peptides: data analysis and applications.” Physical Chemistry Chemical Physics 22.12 (2020): 6492-6506.