Comparative genomics is a powerful tool used in enzyme design and discovery. CD Biosynsis is pleased to support your enzyme discovery and enzyme modification projects with our comparative genomics platform. Here, Whatever your research purpose, our expert team will determine the best solution for your project based on specific requirements.
Overview
A comparative genomics approach, known as genomic context analysis, is one that focuses on identifying associations between genes and proteins in different genomes that may point to functional interactions and suggest the function of unknown proteins. More importantly, this method integrates various types of genomic evidence, such as phylogeny of protein families, domain fusions, gene neighborhoods, expression patterns, metabolic reconstructions, and shared regulatory sites, providing a new method to elucidate gene functions. In the field of enzyme, comparative genomics can provide valuable insights into enzyme design and discovery by facilitating the identification of potential candidate enzymes, understanding enzyme evolution and function, and discovering new enzyme activities. A classic example is the application of comparative genomics to a series of purified proteins from Saccharomyces cerevisiae and the successful identification of several novel enzymes involved in tRNA metabolism in yeast.
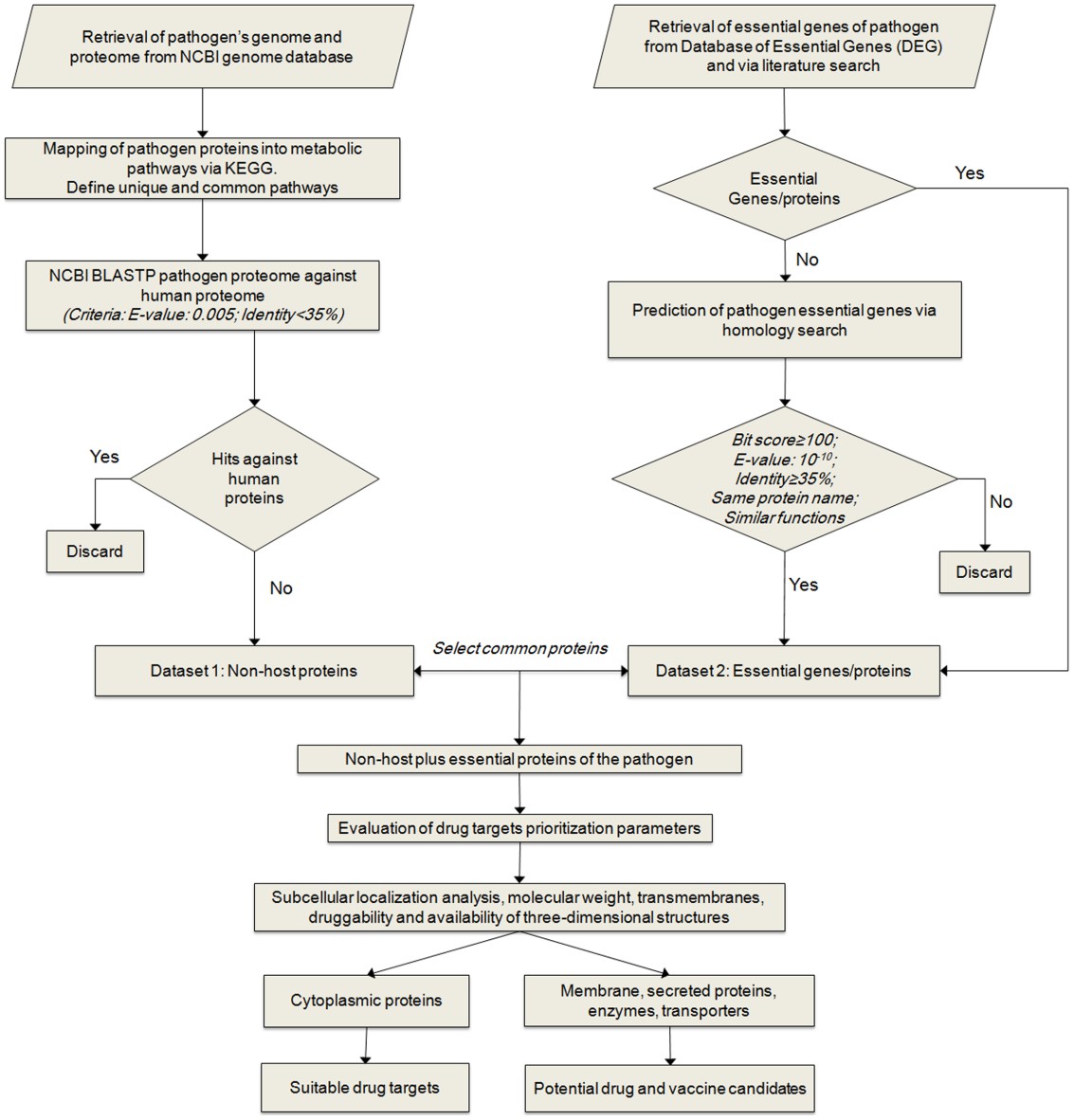 Fig. 1. Comparative genomics workflow. (Butt AM, et al., 2012)
Fig. 1. Comparative genomics workflow. (Butt AM, et al., 2012)
Our Services
Our company has rich experience in the field of using artificial intelligence to promote enzyme evolution. Our expert team uses advanced bioinformatics analysis tools, including genomic analysis, comparative genomic analysis, computer analysis, etc., to discover and design enzymes with specific activities. Here we provide you with first-class comparative genomic analysis services to identify similarities and differences in various organisms. This service strategy mainly involves comparing DNA sequences, genetic structures, and related information of various organisms.
- A general workflow of comparative genomics for enzyme discovery
- Data Mining and Collection
Gaining 16S rRNA gene sequences and annotations of different organisms of interest by accessing publicly available databases or other means, for example, large-scale comparative genomic, pangenomic approaches, metagenomic analysis or metabolic pathway reconstruction.
Use bioinformatics tools such as BLAST or ClustalW to align the genomic sequences of interest. This allows for the identification of genes or gene clusters that show similarities across different organisms.
- Phylogenetic Trees Construction
Construct phylogenetic trees based on the aligned sequences to determine the evolutionary relationships between the organisms. This helps in identifying homologous genes or gene families that are likely to have similar functions.
- Phylogenetic Trees Teconciliation (Optional Step)
We take advantage of the "Reconcile tree" function of PhyML to obtain more reliable information regarding potential horizontal gene transfer events and gene shuffling across taxa.
- Gene Annotation and Functional Prediction
Annotate the identified genes based on their sequence similarities to known enzymes or protein domains. Use functional prediction tools like InterProScan or Pfam to assign putative functions to the genes.
Analyze the expression patterns of the identified genes in different tissues or under different environmental conditions by comparing transcriptome data or using high-throughput techniques like RNA-seq.
Select a subset of candidate genes based on their predicted functions and expression patterns for further experimental validation. Clone and express the genes in a suitable host system and purify the corresponding enzymes. Perform biochemical assays to determine the enzymatic activities and catalytic properties.
In addition, our scientists can use comparative genomics analysis to obtain a large amount of information on the genomes of organisms that produce specific enzymes versus those that do not. This information can help you design or modify enzymes to achieve desired functions or discover novel enzymes with similar or improved properties.
What Can Our Comparative Genomics Platform Resolve?
- Discover novel enzymes with unique functions and potential applications in various industries.
- Understand the enzyme activity you are interested in and its role in biological processes to provide data support for subsequent applications.
- Save time and resources for you by using comparative genomics to prioritize candidate genes for enzyme characterization, rather than screening large libraries or relying solely on experimental approaches.
- Support the discovery of enzyme variants with altered activity or enhanced properties that can be used in a variety of biotechnological applications.
CD Biosynsis aims to provide a powerful analytical tool to accelerate the design of your novel enzyme discovery. Our dedicated team of enzyme experts will customize the best solution for your enzyme application fields. If you are interested in our services or need more detailed information, please feel free to contact us.
Reference
- Butt, AM.; et al. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One. 2012, 7(8):e43080.

































 Fig. 1. Comparative genomics workflow. (Butt AM, et al., 2012)
Fig. 1. Comparative genomics workflow. (Butt AM, et al., 2012)