Computational Modeling of Enzyme-Ligand Interactions is an integral part of modern drug discovery and design processes. By employing sophisticated computer simulations, we can predict how enzymes and ligands (usually potential drug molecules) will interact. This understanding of interactions is pivotal in creating effective therapeutic solutions. This approach doesn't only streamline the development process but also significantly reduces the cost and time required for laboratory experiments, making it a vital tool in the realm of medicinal chemistry and pharmaceutical research.
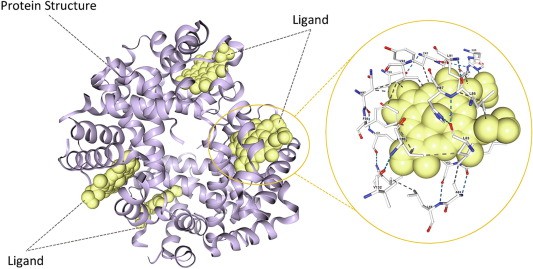 Computational modeling as a tool to Investigate enzyme-ligand binding site (Juan J. Perez, et al., 2021)
Computational modeling as a tool to Investigate enzyme-ligand binding site (Juan J. Perez, et al., 2021)
Technical Route
Our technical route is comprehensive and involves a series of critical stages, each of which has been meticulously planned and designed to ensure the overall success of our operation:
| Stage |
Objective |
Description |
| Molecular Docking |
Predict the orientation of ligands. |
This technique predicts how a ligand binds to a target enzyme, forming a stable complex. It's crucial for initial analysis, providing insight into potential drug interactions. |
| Molecular Dynamics Simulations |
Understand complex evolution |
Simulates physical movements of enzyme-ligand complexes over time, offering a dynamic view of their interaction. This approach provides insights into stability under varying conditions and time scales. |
| Free Energy Calculation Methods |
Estimate binding affinity |
Utilizes free energy calculation methods to estimate the binding affinity between enzymes and ligands. This crucial metric aids in evaluating drug efficiency by gauging the strength of enzyme-ligand interactions. |
Service Process
We offer accurate insights in Computational Modeling of Enzyme-Ligand Interactions via a six-step method. This includes understanding your goals, gathering necessary data, using advanced computational techniques, and delivering detailed findings.
1. Consultation and project planning: Our process kicks off with an in-depth discussion regarding your specific needs and objectives. This helps us to understand your requirements better and enables us to develop a customized project plan that aligns with your goals.
2. Data collection and preparation: Following the initial consultation, we move on to the crucial stage of gathering and preparing the necessary data for computational modeling. This may encompass a range of data including structural data, properties of the ligand, and other information that is pertinent to your project.
3. Molecular docking: The next phase of our process involves performing molecular docking. This scientific method is used to predict the orientation of the ligand when it binds to the target enzyme, which is a key step in understanding how potential drugs interact with their targets.
4. Molecular dynamics simulations: After the docking process, we conduct molecular dynamics simulations. This allows us to gain insights into the potential evolution of the enzyme-ligand complex over time, under various conditions.
5. Analysis of results: The data from the docking and simulations are then meticulously analyzed. Our focus here is on understanding the stability of the complex, the nature of the interactions, and the binding affinity.
6. Report preparation and delivery: Finally, we compile our findings and insights into a comprehensive report. This report, along with any relevant recommendations based on our analysis, is then delivered to you. We're here to assist you. If you have any questions, need more information, or would like to discuss a potential project, please don't hesitate to contact us. Our team is always eager to help and share our expertise.
Applications
| Application |
Description |
| Drug Discovery |
Facilitates drug discovery by computationally modeling enzyme-ligand interactions. This approach aids in virtual screening of compound libraries, guiding the design of enzyme inhibitors and drug leads with improved binding affinity and specificity. |
| Biocatalysis |
Enhances biocatalytic processes by modeling enzyme-ligand interactions, elucidating catalytic mechanisms and substrate binding modes. This enables the rational design and optimization of enzymes for industrial applications such as bioremediation, biofuel production, and pharmaceutical synthesis. |
| Enzyme Engineering |
Supports enzyme engineering endeavors by computationally modeling enzyme-ligand interactions. This assists in understanding structure-function relationships, guiding the design of enzymes with tailored properties for various biotechnological applications. |
| Protein Structure Prediction |
Aids in protein structure prediction by modeling enzyme-ligand interactions, offering insights into protein folding and stability. This facilitates the generation of accurate 3D models for enzymes, enabling structure-based drug design and protein engineering studies. |
| Metabolic Engineering |
Assists in metabolic engineering efforts by modeling enzyme-ligand interactions within metabolic pathways. This helps in the rational design and optimization of metabolic networks for the biosynthesis of valuable compounds such as pharmaceuticals, biofuels, and chemicals. |
| Systems Biology |
Supports systems biology research by modeling enzyme-ligand interactions within cellular networks. This contributes to the understanding of complex biological systems, guiding the elucidation of metabolic and signaling pathways, and aiding in the design of synthetic biology applications. |
FAQs
Here are some frequently asked questions and answers about Computational Modeling of Enzyme-Ligand Interactions. It includes questions about the definition of ligands and enzymes, the importance of understanding enzyme-ligand interactions, the accuracy of computational models, and more. If you don't find the answer you're looking for, feel free to reach out to us.
Q: Why is it important to understand enzyme-ligand interactions?
A: Understanding enzyme-ligand interactions is important because it aids in the design of more effective drugs. It provides insights into how drugs interact with their target enzymes in the body, which can inform the development of drugs with improved efficacy and safety profiles.
Q: How accurate are computational models?
A: While computational models cannot wholly replicate the complexity of biological systems, they can provide valuable insights and predictions about enzyme-ligand interactions. These models are continually refined and validated against experimental data to improve their accuracy.
Q: Can this technique be used for all types of drugs?
A: Computational modeling is versatile and can be used to study interactions for a broad range of potential drug molecules. It can be applied to small molecule drugs, biologics, and even complex multi-component systems.
Q: Is computational modeling a replacement for laboratory experiments?
A: No, computational modeling is not a replacement for laboratory experiments but rather a powerful tool that complements and informs experimental work. It can provide predictions and insights that guide the design of experiments, making the experimental process more efficient.
Q: Can you model interactions involving multiple ligands?
A: Yes, it is possible to model interactions involving multiple ligands. This can be particularly useful in studying drug combinations or understanding the effects of multiple drugs on a single target.
Q: How long does the computational modeling process take?
A: The time required for computational modeling can vary greatly depending on the complexity of the system being studied, the level of detail required, and the computational resources available. However, it is generally much faster than conducting equivalent laboratory experiments.
Q: What are the limitations of computational modeling?
A: While computational modeling is powerful, it does have limitations, including approximations in the models, the need for high computational power for complex systems, and the inability to fully capture the dynamic nature of biological systems.
Q: What kind of data is needed for computational modeling?
A: Data needed for computational modeling usually includes structural data of the target protein or enzyme, properties of the ligand, and sometimes experimental data for validation.
Q: How is the accuracy of computational models validated?
A: The accuracy of computational models is usually validated by comparing the predictions from the model with experimental data.
Q: Can computational modeling predict side effects of drugs?
A: While not its primary purpose, computational modeling can help predict potential side effects by identifying off-target interactions of a drug molecule.
Q: Can computational modeling handle large biomolecules like proteins?
A: Yes, computational modeling can handle large biomolecules, but the complexity and computational demands increase with the size of the molecules.

































 Computational modeling as a tool to Investigate enzyme-ligand binding site (Juan J. Perez, et al., 2021)
Computational modeling as a tool to Investigate enzyme-ligand binding site (Juan J. Perez, et al., 2021)