Our company is a leading provider of cutting-edge enzyme design solutions, specializing in Enzyme Active Site Flexibility Engineering Services. With our expertise in protein engineering and enzymology, we offer innovative and customized approaches to enhance the flexibility and adaptability of enzyme active sites, driving advancements in various industries.
Enzyme active site flexibility engineering refers to the process of modifying the active site of an enzyme to enhance its flexibility or rigidity. The active site of an enzyme is the region where the substrate binds and undergoes a chemical reaction. The flexibility of the active site plays a crucial role in enzyme function and catalytic efficiency.
What We Offer
In the field of enzyme active site flexibility engineering, here are several services can be offered to you for your research.
Consultation and project design
We can provide expert consultation to clients, assisting in project design and development of strategies for enzyme active site flexibility engineering. We can collaborate with clients to define project goals, establish experimental approaches, and develop a comprehensive plan to achieve desired outcomes.
Computational modeling and simulations
Our computational experts can utilize advanced modeling and simulation techniques to predict and analyze the effects of mutations or modifications on enzyme active site flexibility. This service provides valuable insights into the structural dynamics and functional implications of engineered enzymes, aiding in rational design efforts.
Protein engineering and mutagenesis
We can perform targeted mutagenesis or modification of enzymes using techniques such as site-directed mutagenesis or DNA shuffling. Our expertise allows for precise manipulation of enzyme sequences to introduce specific changes in active site flexibility.
High-throughput screening
We can develop and execute high-throughput screening assays to evaluate the activity and flexibility of enzyme variants. Our screening platforms enable rapid screening of large libraries of enzyme variants, facilitating the identification of variants with optimized active site flexibility.
Structural characterization
Our team can provide structural analysis services using techniques such as X-ray crystallography, NMR spectroscopy, or cryo-EM. These methods allow for the determination of the three-dimensional structure of enzymes, including their active sites, providing critical insights into the relationship between structure, flexibility, and function.
Enzyme optimization and evolution
We can perform directed evolution experiments to optimize enzyme variants with desired active site flexibility characteristics. Our iterative optimization process, combined with high-throughput screening, allows for the identification of enzyme variants with improved flexibility and catalytic performance.
Pharmacokinetic and pharmacodynamic studies
We can conduct preclinical studies to assess the pharmacokinetic and pharmacodynamic properties of engineered enzymes. This includes evaluating enzyme stability, bioavailability, clearance, and therapeutic efficacy in relevant preclinical models.
Safety and toxicity assessments
We can perform safety and toxicity assessments of engineered enzymes to evaluate their potential adverse effects. This includes conducting in vitro and in vivo studies to assess the safety profile, immunogenicity, and potential off-target effects of the engineered enzymes.
Data analysis and reporting
We offer comprehensive data analysis and reporting services to interpret and present experimental results. Our reports provide clear and concise summaries of the findings, facilitating informed decision-making and further development.
By offering these services, we aim to support our clients in their enzyme active site flexibility engineering projects, providing specialized expertise and preclinical evaluation to advance their development efforts. Please contact us and start our cooperation.
Reference
- Nestl, B.M.; B, Hauer. Engineering of Flexible Loops in Enzymes. ACS Catalysis, 2014. 4(9): p. 3201-3211.

































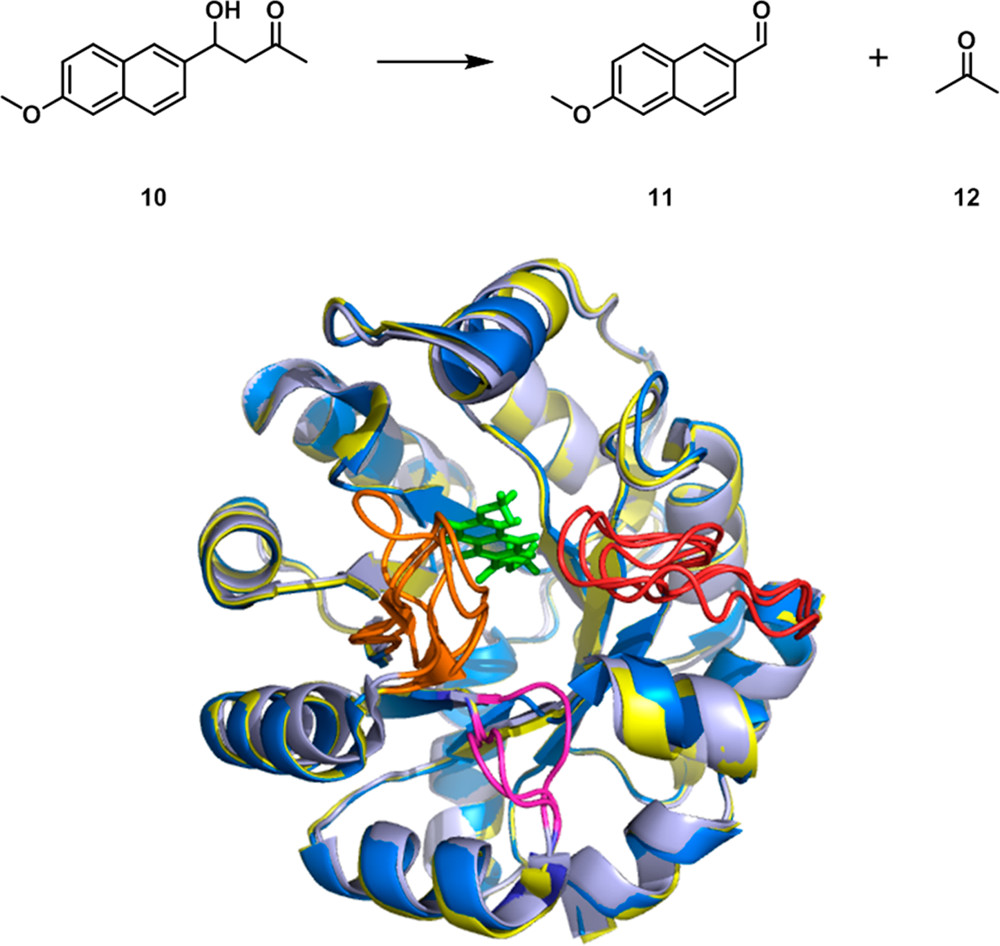 Fig. 1. Loop Mobility and Active Site Remodeling. (Nestl, B.M.; B, Hauer, 2014)
Fig. 1. Loop Mobility and Active Site Remodeling. (Nestl, B.M.; B, Hauer, 2014)