Enzymes Bioinformatics Analysis is a field of study that combines bioinformatics and enzymology to analyze and interpret the information related to enzymes. It utilizes computational tools and methods to gather, process, and analyze data about enzyme sequences, structures, functions, and interactions. The use of Enzymes Bioinformatics Analysis is critical due to its vast applications in various fields. It can aid in the discovery of new drugs, contribute to the diagnosis of diseases, help in the creation of genetically improved crops, and assist in understanding ecological systems. Furthermore, it can provide valuable insights from the data that can be used for further research and studies.
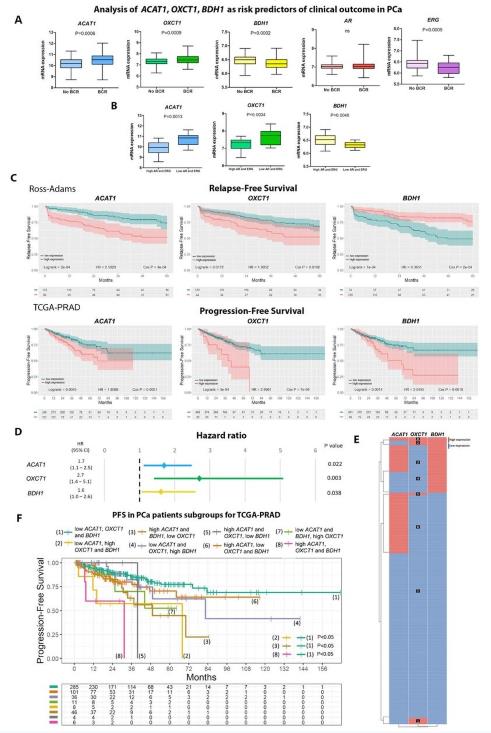 Bioinformatics analysis of ketogenic/ketolytic enzymes (Estefania L., et al., 2021)
Bioinformatics analysis of ketogenic/ketolytic enzymes (Estefania L., et al., 2021)
Service Process
Our AI Guided Precise HTP Library Screening process is designed to leverage the power of artificial intelligence in the screening of libraries, providing a more efficient, precise, and cost-effective solution. This meticulous and advanced approach reduces potential human error and enables the identification of intricate patterns and relationships that may not be evident through traditional methods. The process involves six key steps:
- Define project scope and requirements: In the initial phase, we establish a close collaboration with our clients in order to gain a comprehensive understanding of their specific requirements and the overall scope of the project. This step is vital for ensuring that our services align with the client's goals and expectations.
- Data collection: Next, we embark on the task of gathering all relevant data that is essential for the high-throughput screening (HTS) process. This involves identifying the right data sources, ensuring the quality of data, and preparing it for further processing.
- AI model training: Once we have collected the necessary data, we proceed to train the Artificial Intelligence (AI) model. Utilizing the amassed data, the AI model is trained to comprehend and adapt to the unique needs of the project, thereby enabling accurate prediction and analysis capabilities.
- Predictive screening: Armed with a trained AI model, we guide the high-throughput screening process, making accurate predictions which significantly increase the efficiency of the screening process.
- Optimization and result analysis: Post the predictive screening, we analyze the results and use these insights to optimize both the AI model and the HTS process. This step ensures that we consistently improve our processes, enhancing the accuracy of our predictions and the effectiveness of our screening.
- Report generation and delivery: Upon completion of the screening process, we compile the results into detailed reports. These comprehensive documents are then delivered to our clients, providing them with a thorough understanding of the results and the steps taken to arrive at these conclusions.
We're here to assist you. If you have any questions, need more information, or would like to discuss a potential project, please don't hesitate to contact us. Our team is always eager to help and share our expertise.
Applications
| Method |
Application |
Description |
| Sequence Alignment |
Drug Discovery |
Utilizes sequence alignment algorithms to compare enzyme sequences, facilitating drug target identification and lead compound optimization. This method helps identify conserved regions and functional domains critical for enzyme activity, aiding in the rational design of enzyme inhibitors and therapeutics. |
| Structural Bioinformatics |
Biocatalyst Engineering |
Applies structural bioinformatics tools to analyze enzyme structures, elucidating catalytic mechanisms and substrate binding interactions. By integrating structural data with computational simulations, this approach enables the rational engineering of enzymes for enhanced biocatalytic performance in industrial processes, such as bioremediation, biofuel production, and pharmaceutical synthesis. |
| Functional Annotation |
Protein Engineering |
Employs functional annotation techniques to annotate enzyme functions and predict substrate specificity. By leveraging genomic and proteomic data, this method facilitates the identification of enzymes with desired properties for various biotechnological applications, including enzyme optimization, pathway engineering, and metabolic engineering. |
| Molecular Docking |
Drug Design |
Utilizes molecular docking algorithms to predict enzyme-ligand interactions and evaluate potential drug candidates. This computational approach aids in virtual screening of compound libraries, guiding the design of enzyme inhibitors and drug leads with improved binding affinity and specificity. |
| Network Analysis |
Systems Biology |
Applies network analysis methods to study enzyme interaction networks and metabolic pathways. By analyzing enzyme connectivity and pathway topology, this approach provides insights into cellular metabolism and regulatory mechanisms, guiding the rational engineering of metabolic pathways for bioproduction of chemicals, pharmaceuticals, and biofuels. |
| Homology Modeling |
Structural Biology |
Utilizes homology modeling techniques to predict enzyme structures based on homologous templates. This method enables the generation of 3D models for enzymes with unknown structures, facilitating structure-based drug design, protein engineering, and molecular simulations to study enzyme dynamics and function. |
| Machine Learning |
Enzyme Function Prediction |
Utilizes machine learning algorithms to predict enzyme functions and substrate specificities based on sequence and structural features. By training predictive models on large-scale enzyme datasets, this approach enables accurate annotation of enzyme functions and guides the discovery of novel enzymes with desired properties for biotechnological applications. |
Case Study
The figure below is a case of bioinformatics analysis of enzymes involved in the tricarboxylic acid cycle: (A) The enzyme shown are Pyruvate dehydrogenase E1 component subunit alpha (PDH-E1I), Citrate synthase (CISY), isocitrate dehydrogenase (IDH), succinyl-CoA ligase subunit beta (SUCB), succinate dehydrogenase (DHSA), fumarate hydratase (FH), malate dehydrogenase (MDH). The triangle present acetylation sites and the circle present succinylation sites. Red present up-regulated modification and blue present the oppsite. The oval present mitochondrial and the rectangle present cytoplasm. (B) Clustal W alignment of isocitrate dehydrogenase NADP (mitochondrial) homologs from H. sapiens (GI:583966148), M. musculus (GI:225579033), S. cerevisiae (GI:124160), A. thaliana (GI:75246494). Conserved succinyllysine residues are labelled with arrows. (C) Isocitrate dehydrogenase NAD subunit beta (mitochondrial) homologs from H. sapiens (GI:385648280), M. musculus (GI:18700024), S. cerevisiae (GI:6324291), A. thaliana (GI:330251495). Conserved succinyllysine site are labelled with arrows.
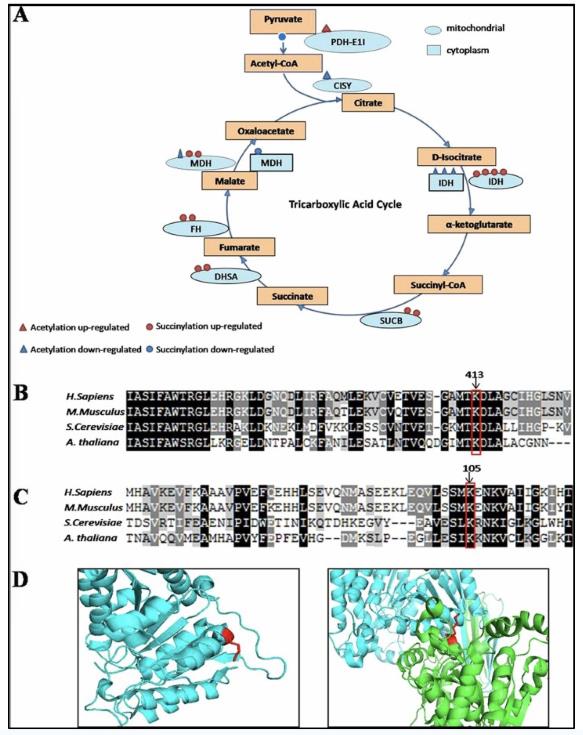 Bioinformatics analysis of enzymes involved in the tricarboxylic acid cycle (Hong Xu, et al., 2016)
Bioinformatics analysis of enzymes involved in the tricarboxylic acid cycle (Hong Xu, et al., 2016)
FAQs
To help you better understand our Enzymes Bioinformatics Analysis service, we have compiled a list of some frequently asked questions. These cover a range of topics from the purpose and applications of the service to its technical aspects, data security, and the support we provide to our clients. If you don't find the answer you're looking for, feel free to reach out to us.
Q: Could you explain the purpose of Enzymes Bioinformatics Analysis in more detail?
A: The primary purpose of Enzymes Bioinformatics Analysis is to delve deeper into the intricate understanding of the functions and structures of enzymes. This understanding can be pivotal in various fields, including drug discovery, disease modeling, and more.
Q: Who are the targeted users of this service?
A: Our service is designed to be utilized by a broad range of professionals. This includes researchers, scientists, and businesses operating in the field of biotechnology. Essentially, anyone looking to gain insights into the role of enzymes can benefit from this service.
Q: Can you assure the security of the data used?
A: Absolutely, we place the utmost importance on the security of your data. We adhere to stringent security protocols to make sure your data is handled with the highest level of protection.
Q: What is the expected turnaround time for the analysis?
A: The turnaround time for our Enzymes Bioinformatics Analysis is largely dependent on the complexity and size of the project. However, we do our best to provide results in a timely and efficient manner.
Q: What types of data formats are accepted for the analysis?
A: We strive to accommodate a variety of needs and thus accept data in various formats. This includes but is not limited to TXT, CSV, and more.
Q: Is there any support provided after the analysis has been completed?
A: Yes, in addition to providing a comprehensive analysis, we also offer post-analysis support to help you interpret the results and apply them effectively.
Q: Is it possible to get a personalized analysis?
A: Certainly, we understand that each project has unique needs. Therefore, we offer to customize the analysis based on your specific requirements and research goals.
Q: What bioinformatics software is utilized for the analysis?
A: Our team uses a variety of cutting-edge software tools for the analysis. This includes widely recognized tools such as BLAST, FASTA, among others.
Q: How reliable and accurate is the analysis?
A: We pride ourselves on the quality of our work. Our team of experts strives for the highest accuracy in our analysis, using rigorous methods and the latest bioinformatics tools.

































 Bioinformatics analysis of ketogenic/ketolytic enzymes (Estefania L., et al., 2021)
Bioinformatics analysis of ketogenic/ketolytic enzymes (Estefania L., et al., 2021) Bioinformatics analysis of enzymes involved in the tricarboxylic acid cycle (Hong Xu, et al., 2016)
Bioinformatics analysis of enzymes involved in the tricarboxylic acid cycle (Hong Xu, et al., 2016)