Evolutionary Trace Analysis is a cutting-edge method that allows us to identify the most crucial functional regions in proteins by tracking their evolutionary modifications over time. This technique is extraordinarily beneficial for enzymes, as it allows us to focus on the active sites and essential residues that play a significant role in their catalytic activities. By doing so, we can understand the fundamental characteristics that contribute to an enzyme's function and how it may be manipulated for various applications.
 Bypassing evolutionary dead ends and switching the rate-limiting step of a human immunotherapeutic enzyme (John Blazeck, et al., 2022)
Bypassing evolutionary dead ends and switching the rate-limiting step of a human immunotherapeutic enzyme (John Blazeck, et al., 2022)
Technical Route
Our technical route is a rigorous and comprehensive process that ensures highly accurate and insightful results that can be instrumental for further research and understanding. This involves a series of meticulously executed steps:
| Steps |
Objective |
Description |
| Collection and alignment of homologous protein sequences |
Gather and align similar protein sequences |
This step involves collecting protein sequences showing similarity and aligning them meticulously. By identifying similarities and differences, it aids in comparative study and understanding sequence relationships. |
| Phylogenetic tree construction |
Illustrate evolutionary relationships |
Construct a phylogenetic tree to visualize evolutionary relationships among proteins. This graphical representation depicts their evolution in relation to each other and their common ancestors. |
| Identification of evolutionary conserved residues |
Identify unchanged residues throughout evolution |
Identify residues that have remained unchanged over time, indicating their functional importance. This step provides insights into key functional areas crucial for protein function. |
| Mapping of these residues onto the 3D structure of the enzyme |
Project residues onto the enzyme's 3D structure |
Project identified crucial residues onto the enzyme's 3D structure for a comprehensive visual representation. This mapping offers insights into the protein's spatial configuration and critical functional areas. |
Service Process
Our service process is designed for a smooth experience delivering tangible results:
1. Initial consultation: We begin with a detailed discussion to understand your specific needs, objectives, and expectations, setting the stage for tailored analysis.
2. Collection of enzyme sequences: We gather necessary enzyme sequences, forming the backbone of our analysis.
3. Performing the Evolutionary Trace Analysis: Our team of experts performs an accurate and insightful analysis using advanced techniques and tools.
4. Detailed report generation: We translate insights into a comprehensive report detailing the results, implications, and their effective application. The value of our analysis lies in the accuracy of our results and our ability to help you apply these findings.
We're here to assist you. If you have any questions, need more information, or would like to discuss a potential project, please don't hesitate to contact us. Our team is always eager to help and share our expertise.
Applications
| Application |
Description |
| Protein Function Prediction |
Evolutionary Trace Analysis is extensively used in predicting protein function by identifying evolutionarily conserved residues critical for structural integrity and functional activity. By analyzing sequence conservation patterns across homologous proteins, Evolutionary Trace Analysis helps decipher the functional importance of specific amino acids, providing insights into key functional regions and biochemical activities of proteins. This information is crucial for understanding protein structure-function relationships, annotating protein function, and guiding experimental studies. |
| Protein Engineering |
Evolutionary Trace Analysis plays a pivotal role in protein engineering, enabling the design and modification of proteins with improved stability, specificity, and catalytic efficiency. By analyzing evolutionary signatures and identifying conserved residues, Evolutionary Trace Analysis guides the rational design of protein variants tailored for specific applications. This approach facilitates the optimization of protein properties, such as substrate specificity, enzymatic activity, and protein-protein interactions, leading to the development of novel enzymes, therapeutic proteins, and biotechnological tools. Protein engineers leverage Evolutionary Trace Analysis to explore sequence-function relationships, predict mutational effects, and engineer proteins for various industrial, medical, and biotechnological applications. |
| Drug Target Identification |
Evolutionary Trace Analysis is a valuable tool for drug target identification, aiding in the discovery and validation of potential drug targets for therapeutic intervention. By analyzing evolutionary conservation patterns in protein sequences, Evolutionary Trace Analysis identifies conserved residues essential for protein function and stability, providing valuable insights into the druggability of protein targets. This approach facilitates the prioritization of candidate drug targets based on their evolutionary conservation, functional significance, and potential for therapeutic modulation. Drug discovery researchers leverage Evolutionary Trace Analysis to screen and prioritize protein targets, design target-specific inhibitors, and optimize therapeutic strategies for various diseases, including cancer, infectious diseases, and neurological disorders. |
| Disease Mechanism Investigation |
Evolutionary Trace Analysis is widely employed in investigating disease mechanisms, offering valuable insights into the molecular basis of disease pathogenesis and progression. By analyzing evolutionary signatures in disease-associated proteins, Evolutionary Trace Analysis identifies conserved residues implicated in pathological pathways, protein-protein interactions, and post-translational modifications. This approach facilitates the identification of key molecular determinants driving disease phenotypes, providing a deeper understanding of disease mechanisms and potential therapeutic targets. Disease researchers utilize Evolutionary Trace Analysis to explore genotype-phenotype correlations, predict the functional impact of disease-associated mutations, and prioritize therapeutic interventions for precision medicine approaches. |
| Structural Biology |
In structural biology, Evolutionary Trace Analysis serves as a powerful tool for elucidating protein structure-function relationships and guiding experimental studies. By analyzing evolutionary conservation patterns in protein sequences, Evolutionary Trace Analysis identifies conserved residues crucial for maintaining structural stability, ligand binding, and catalytic activity. This information is invaluable for protein structure prediction, homology modeling, and structural annotation, providing structural insights into protein function and dynamics. Structural biologists leverage Evolutionary Trace Analysis to predict protein interaction interfaces, rationalize protein-ligand interactions, and engineer protein variants with desired structural properties. This approach facilitates the interpretation of experimental data, the design of functional experiments, and the generation of testable hypotheses in structural biology research. |
| Molecular Evolution Studies |
Evolutionary Trace Analysis is extensively utilized in molecular evolution studies to trace the evolutionary history of proteins and infer functional divergence across species and evolutionary time. By analyzing evolutionary conservation patterns in protein sequences, Evolutionary Trace Analysis identifies conserved residues shared among homologous proteins, providing insights into their evolutionary relationships and functional constraints. This approach enables researchers to reconstruct evolutionary pathways, infer ancestral protein sequences, and investigate the adaptive evolution of protein function. Molecular evolutionists leverage Evolutionary Trace Analysis to study protein evolution dynamics, predict evolutionary trajectories, and explore the origins of biological diversity across taxa. This information enhances our understanding of evolutionary processes, functional adaptation, and the molecular basis of phenotypic diversity in living organisms. |
Case Study
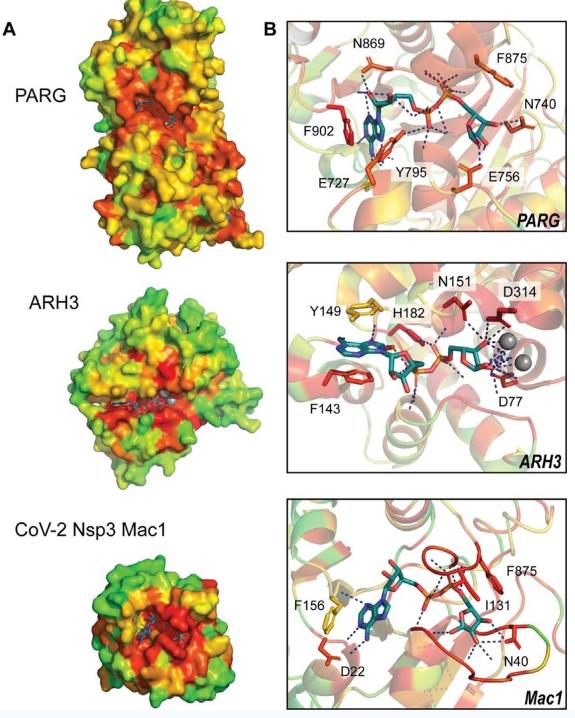
The figure illustrates how Evolutionary Trace Analysis pinpoints functional residues within the active sites of PARG, ARH3, and CoV-2 Nsp3 Mac1. High-ranking ET values (red-orange) cluster at the active sites of each enzyme. Inspection of residues within the active sites (right panel) shows greatest conservation among residues coordinating the ADPr pyrophosphate linker and terminal ribose and increased variation within the adenine pocket.
 Constructs of yCD variants and their relative activity (Davide Moiani, et al., 2021)
Constructs of yCD variants and their relative activity (Davide Moiani, et al., 2021)
FAQs
We understand that Evolutionary Trace Analysis (ETA) might raise numerous questions, given its technical nature and wide-ranging applications. Thus, we've made a comprehensive list of inquiries and their answers to ensure you have a deep understanding of this critical tool:
Q: Can ETA be applied to enzymes?
A: Yes, ETA can be applied to enzymes. It can help in identifying key functional residues that are crucial for enzymatic activity. By identifying key functional residues in an enzyme, researchers can potentially modify these residues to alter the enzyme's function, paving the way for engineered enzymes with desired characteristics.
Q: Can ETA help in the design of enzyme inhibitors?
A: Yes, by identifying key functional residues in enzymes, ETA can guide the design of inhibitors that target these residues, which can potentially lead to more effective therapeutic agents.
Q: Is this technique useful for all types of enzymes?
A: Absolutely, Evolutionary Trace Analysis is versatile and can be applied to all types of enzymes, providing valuable insights regardless of the enzyme's specific function or structure.
Q: What are the main applications of ETA?
A: ETA is a versatile tool with a wide range of applications. Its primary uses include drug design, understanding disease mechanisms, and protein engineering. It's an essential instrument for both academic research and industrial applications, including biotechnology and pharmaceuticals.
Q: What type of information does ETA provide?
A: ETA provides vital information on the key functional residues in a protein. These functional residues are crucial to how the protein works. By understanding these residues, researchers can gain insights into how the protein functions and how it might be modified for therapeutic purposes.
Q: How does ETA contribute to drug design?
A: ETA identifies the key functional residues in proteins. This information can guide the design of drugs that target these functional residues, potentially leading to more effective therapeutics.
A: While no method is 100% accurate, ETA is a robust tool that has been widely validated. The accuracy of ETA largely depends on the quality and amount of the homologous sequences available for study.
Q: What resources are needed to perform ETA?
A: To perform ETA, you need sequences of homologous proteins and computational resources to align these sequences, create a phylogenetic tree, and perform the trace analysis.

































 Bypassing evolutionary dead ends and switching the rate-limiting step of a human immunotherapeutic enzyme (John Blazeck, et al., 2022)
Bypassing evolutionary dead ends and switching the rate-limiting step of a human immunotherapeutic enzyme (John Blazeck, et al., 2022) Constructs of yCD variants and their relative activity (Davide Moiani, et al., 2021)
Constructs of yCD variants and their relative activity (Davide Moiani, et al., 2021)