Our company is a pioneer in harnessing the power of artificial intelligence to revolutionize enzyme design and optimization. Our goal is to provide ideal biocatalysts to industries seeking innovative solutions and enhanced enzymatic processes through state-of-the-art in silico analysis services.
Overview
In silico analysis, a valuable tool in enzyme discovery, allows researchers to analyze large datasets of genomic and proteomic information to identify potential enzymes, which provides new methods for the expansion of available genomic databases. For example, in silico analysis can be used to search for gene sequences that encode proteins with known enzymatic activities or to predict the function of uncharacterized proteins based on their sequence and structure. Furthermore, computational analysis methods can be applied to predict the biochemical properties and catalytic mechanisms of enzymes, which can provide insights into their potential applications.
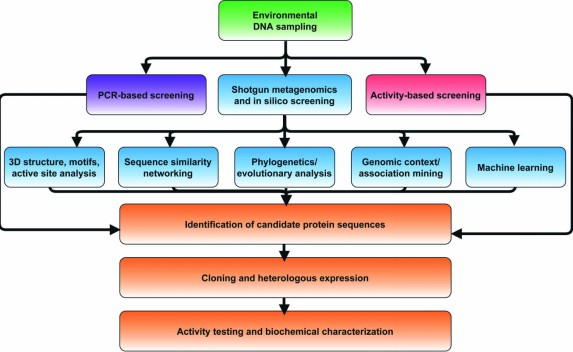 Fig. 1. Flowchart of strategies for in silico selection and experimental characterization of candidate metagenomic enzymes. (Robinson SL, et al., 2021)
Fig. 1. Flowchart of strategies for in silico selection and experimental characterization of candidate metagenomic enzymes. (Robinson SL, et al., 2021)
Our Services
Identifying new enzymes from a large number of candidate reactions with the help of genome mining requires performing downstream bioinformatics steps to make metagenomics publicly available in public repositories. We have established a comprehensive computer analysis platform to assist you in predicting the function of new enzymes in protein families. Here, we provide different in silico analysis methods for enzyme discovery, including but not limited to:
- Phylogenetics Analysis
- Sequence Similarity Network Analysis
- Genetic Context and Interactions
- 3D Structure-Based Analysis
- Motif and Active Site Residue Analysis
- Machine Learning-Based Analysis
Additionally, Our experienced scientists will conduct an in silico evaluation based on the characteristics of your specific enzyme to help you understand the transport mechanisms of molecules in the tunnels present in your enzymatic structures for wider application.
Explore Our General Strategy In Silico Evaluation
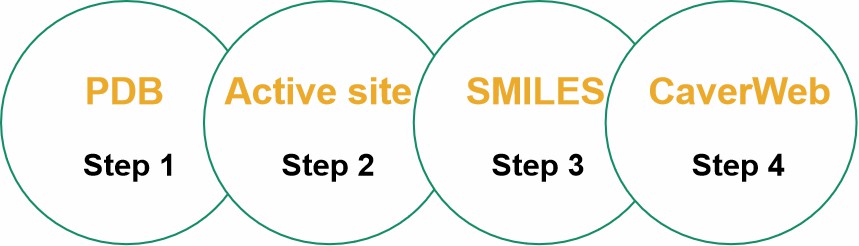 Fig. 2. Our representative scheme of the steps that were performed in the computational analysis.
Fig. 2. Our representative scheme of the steps that were performed in the computational analysis.
- Obtaining the PDB's of the analyzed enzymes in the Protein Data Bank (PDB).
- Structuring and identifying the enzymatic structure as well as identifying the active site of each enzyme.
- Search and selection of the binders and products (SMILES codes) of this analysis.
- Inserting the SMILES codes of the substances into the analyzed enzymes using the CaverWeb server.
Integrating our in silico analysis services throughout your program can accelerate the discovery of enzymes' chances of success. Whether it is enzyme design, enzyme discovery, or using it as a stand-alone service, we can meet your needs.
Service Highlights
- Gain insights into the evolution of functional relationships between enzymes within protein families by ancestral sequence reconstruction.
- Allow users to quickly identify clusters without known representatives in sequence space.
- Facilitate identification of novel targets for enzyme discovery that a unusual coexisting domains or interacting proteins.
- Reveal new functional relationships for proteins independent of primary sequence.
- Identification of enzymes with significant impact on biocatalysis based on changes in active site structure.
- Recognize patterns in big metagenomic datasets to accelerate enzyme discovery.
With our extensive experience and advanced in silico platform, We are confident in offering the best enzyme discovery and design services for our global customers. We guarantee the most satisfactory results helping to accelerate your project development process. Please contact us now for your exclusive solution.
References
- Robinson SL, et al. A roadmap for metagenomic enzyme discovery. Nat Prod Rep. 2021,38(11):1994-2023.
- Azevedo, TSM, et al. In Silico Evaluation of Enzymatic Tunnels in the Biotransformation of α-Tocopherol Esters. Front Bioeng Biotechnol. 2022, 9:805059.

































 Fig. 1. Flowchart of strategies for in silico selection and experimental characterization of candidate metagenomic enzymes. (Robinson SL, et al., 2021)
Fig. 1. Flowchart of strategies for in silico selection and experimental characterization of candidate metagenomic enzymes. (Robinson SL, et al., 2021) Fig. 2. Our representative scheme of the steps that were performed in the computational analysis.
Fig. 2. Our representative scheme of the steps that were performed in the computational analysis.