Many diverse enzymes excavated from metagenomes were widely used in agriculture, food, health, and environmental fields. CD Biosynsis is pleased to support your enzyme discovery and enzyme modification projects with our metagenomics and advanced bioinformatics algorithms. Here, Whatever your research purpose, our expert team will determine the best solution for your project based on specific requirements.
Overview
The application prospects of enzymes and catalysts in biotechnology are very broad. Unfortunately, many microorganisms remain unnamed and cannot be grown in growth media. However, in metagenomics, DNA is extracted directly from environmental samples without going through a laboratory culture process, which provides new ideas for exploring a large number of new enzymes that were hitherto unknown. Metagenomics is a method that directly analyzes the genomes of microbial communities in the environment. A representative and thorough result is obtained when the diversity of bacteria is analyzed using DNA. This allows researchers to acquire data on microbial diversity, with a 99% success rate, and different kinds of genes encode an enzyme that has yet to be found. A typical example is α-xylosidase discovery and, which involved the metagenomic DNA was first extracted from the soil sample. A metagenomic library was constructed and transformed into E. coli, followed by screening for activity.
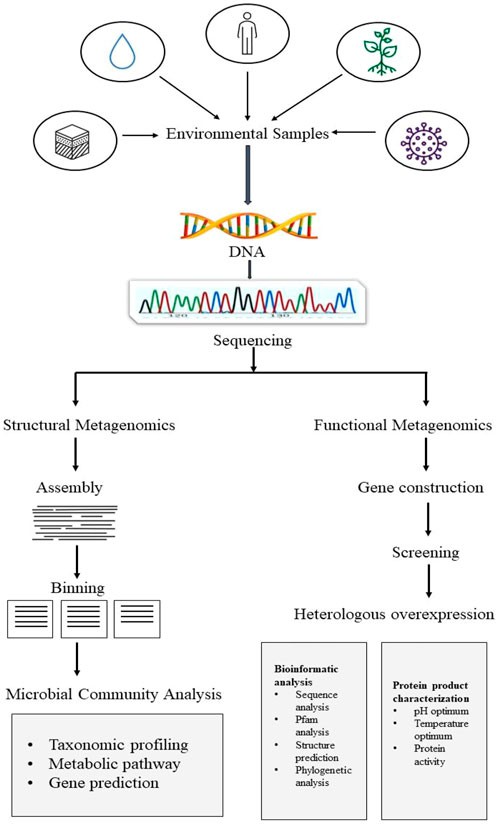 Fig. 1. Framework for metagenomics inculcates its two main studies. (Butt AM, et al., 2012)
Fig. 1. Framework for metagenomics inculcates its two main studies. (Butt AM, et al., 2012)
Our Services
CD Biosynsis is a leading AI enzyme expert with extensive experience in using artificial intelligence to promote enzyme discovery and design. Our expert team is adept at using metagenomics to mine microbial samples from rare or unculturable sources. This service relies heavily on our advanced bioinformatics algorithms and comprehensive data analysis platform.
- One-stop Service for Metagenomic Enzyme Mining
Metagenomics to discover new genes and enzymes often involves metagenomic DNA extraction, metagenomic library construction, heterologous expression, and high-throughput screening. With solid expertise and rich experience in enzyme discovery, our company provides you with professional one-stop services:
- Metagenomics DNA extraction and library construction.
- Genomic database comparison and knowledge-based prediction.
- High-throughput screening.
- Enzyme expression, purification, and characterization.
How do We Discover Novel Enzymes?
Our company utilizes information from metagenomic DNA databases to target the desired genes through various strategies, then validate and characterize them to discover new enzymes. These strategies include:
- Shotgun metagenomics strategy
- Activity-guided screening strategy
- PCR-based screening strategy
Briefly, our molecular modeling tools and advanced bioinformatics algorithms were used firstly to sample and filter the sequence space of the unique metagenome database to construct the final highly specific and selective enzyme set. Then the selected panel is cloned, expressed, and screened for responses of interest.
Support Services
In our company, a variety of online tools, stand-alone software, and servers are used to assess microbial diversity, network analysis, metabolic pathway mapping and functional analysis, including but not limited to:

- MG-RAST
- EBI
- RDP
- QIIME
- MOTHUR
- SqueezeMeta
CD Biosynsis aims to help global customers discover enzymes with known activity but uncommon properties and/or unknown activity on specific substrates through metagenomics and advanced bioinformatics algorithms. Our dedicated team of enzyme experts will customize the best solution for your enzyme application fields. If you are interested in our services or need more detailed information, please feel free to contact us.
References
- Butt, AM.; et al. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One. 2012, 7(8):e43080.
- Robinson, SL.; et al. A roadmap for metagenomic enzyme discovery. Nat Prod Rep. 2023, 38(11):1994-2023.
- Varaljay, VA.; et al. Editorial: Functional Metagenomics for Enzyme Discovery. Front Microbiol. 2022, 13:956106.
- Prayogo, F.A.; et al. Metagenomic applications in exploration and development of novel enzymes from nature: a review. J Genet Eng Biotechnol. 2020, 18(1):39.

































 Fig. 1. Framework for metagenomics inculcates its two main studies. (Butt AM, et al., 2012)
Fig. 1. Framework for metagenomics inculcates its two main studies. (Butt AM, et al., 2012)