CD Biosynsis is a leading expert in the field of enzyme design and evolution, with a long and in-depth experience in enzyme directed evolution. Our dedicated team utilizes evolutionary trace analysis to extract key information from enzyme sequences, providing valuable insights for the rational design of enzymes with improved properties or novel functions.
Overview
Evolutionary trace is a method for identifying functionally important residues in proteins. It closely links the sequence variation to the evolutionary differences, as the differences between different sequences are matched to their functional differences. In other words, evolutionary trace is actually a quantitative test for sequence differences versus functional differences. The functionally important residues in proteins are often conserved during evolution, so we are able to leverage the evolutionary trace method, which allows us to analyze a diverse set of related protein sequences to identify their conserved sequences and predict the functional significance of specific residues.
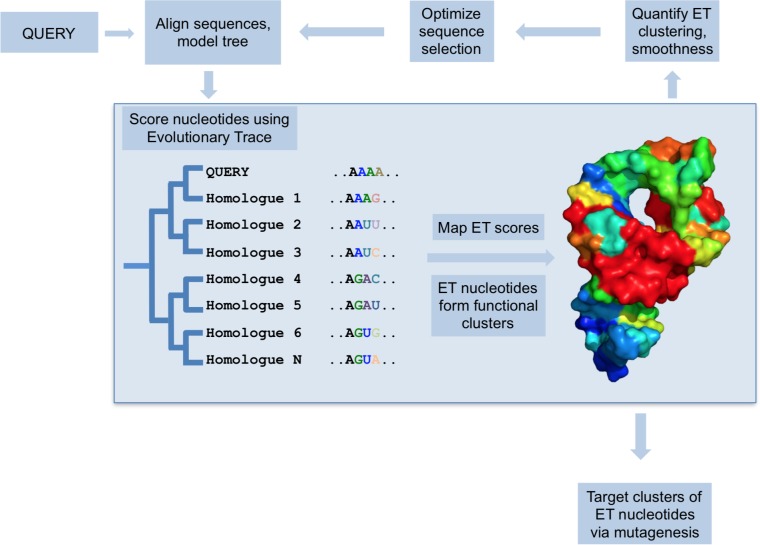 Fig 1. The evolutionary trace model. (Novikov IB, et al., 2020)
Fig 1. The evolutionary trace model. (Novikov IB, et al., 2020)
Our Services
Based on the evolutionary trace method, our EnzymoGenius™ platform provides prediction and identification services of active sites or key catalytic amino acid residues of enzymes, enabling researchers to better carry out enzyme engineering studies and applications.
- Sequence Alignment
First of all, we can sequence the amino acid sequences of the target enzyme. After sequencing, we search for proteins that are highly homologous to its amino acid sequence, and compare and analyze their sequences to find out the conserved and variable regions in the amino acid sequence.
- Evolutionary Trace Calculation
After sequence evaluation, an evolutionary trail score was assigned to different amino acid residues based on their conservation in the sequence comparison results. Highly conserved amino acid residues tend to have higher scores, indicating a potentially important function.
- Mapping to Enzyme Structure
If the target enzyme has a corresponding 3D structure, the evolutionary trace score can be simultaneously mapped to that structure.
- Identification of Critical Residues
Amino acid residues with high evolutionary trace scores are generally considered to be critical residues, indicating that they play an essential role in protein structure or function.
Our Advantages
- Professional Scientific Team
We have a professional research team of scientists with decades of experience in the field of enzyme design and evolution, focusing on the directed evolution and rational design of enzymes, and are committed to providing the greatest impetus for enzyme biology research.
- AI-assisted Development and Design
Combined with our advantage in AI applications, our EnzymoGenius™ platform enables the application of AI-assisted modeling to evolutionary trace methods, resulting in a powerful increase in predictive efficiency.
- High-quality Services
We have a team of highly skilled and experienced scientists with in-depth knowledge of enzyme design evolution, who can provide you with professional consulting services before the project and reliable support services after the sale.
CD Biosynsis strives to offer comprehensive and reliable enzyme evolution trace analysis for various applications, whether academic research, industrial applications, or biotechnological development, our services are tailored to meet your specific requirements. Contact us today to learn more about our enzyme evolutionary trace analysis services and how they can benefit your research or development projects. Our team is ready to assist you and provide the insights you need to achieve your goals.
Reference
- Novikov, IB.; et al. An Evolutionary Trace method defines functionally important bases and sites common to RNA families. PLoS Comput Biol. 2020, 16(3):e1007583.

































 Fig 1. The evolutionary trace model. (Novikov IB, et al., 2020)
Fig 1. The evolutionary trace model. (Novikov IB, et al., 2020)