Predicting Substrate Binding Pathways through Simulation stands as an innovative computational modeling technique at the forefront of modern science. It employs state-of-the-art algorithms designed to accurately predict the way substrates, which can be molecules or ions, bind to specific sites within a biological system. This technique is not only instrumental in advancing our understanding of complex biological processes, but it is also key to the development of new drugs and the improvement of biochemical processes.
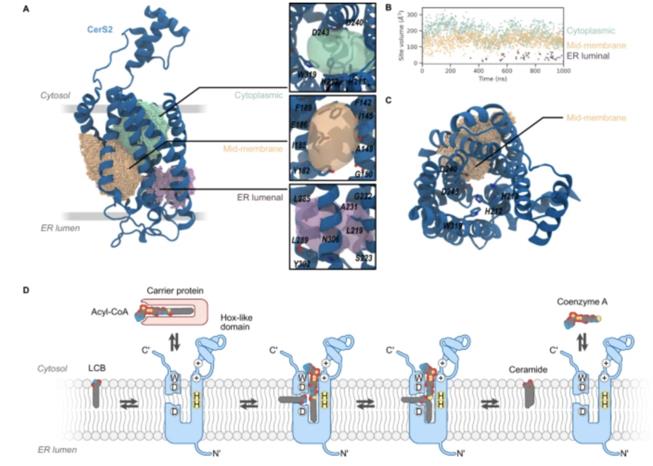 Computational design and molecular dynamics simulations suggest the mode of substrate binding in ceramide synthases (Iris D. Zelnik, et al., 2023)
Computational design and molecular dynamics simulations suggest the mode of substrate binding in ceramide synthases (Iris D. Zelnik, et al., 2023)
Technical Route
Our technical approach is defined by using a range of advanced techniques and methodologies. Key among these are molecular dynamics simulations, which let us observe and predict how molecules behave in a dynamic system. We also use Monte Carlo sampling to explore the probability distribution of potential outcomes, providing a strong statistical basis for our investigations. Additionally, we utilize machine learning algorithms to learn from data and make predictions or decisions. These algorithms are useful in spotting patterns and trends that might otherwise be hard to see. By integrating these advanced methods, we can do more than just analyze - we can accurately predict complex processes like substrate binding, thereby improving our overall knowledge and ability in the field.
Service Process
Our procedure for predicting substrate binding pathways through simulation includes several critical steps, each contributing to the final, precise results. Here's an outline of our approach:
1. Initial Consultation: In this crucial first step, we engage in an in-depth discussion with you. We aim to understand your specific needs, objectives, and the details of the biological system you want to model. Our goal is to fully comprehend your requirements to deliver the most accurate results.
2. Data Collection: Once we have a clear understanding of your needs, we begin data collection. We gather all necessary data for the modeling process, including the structural data of the biological system, substrate details, and any other relevant information.
3. Model Building: Using the collected data, our team applies advanced computational techniques to construct a detailed and comprehensive model. This model simulates the substrate binding process, accurately representing the biological system you're studying.
4. Simulation: After rigorously testing the model for accuracy, we run the simulation. Throughout this phase, we observe the process, record necessary data, and analyze the results in real-time.
5. Report: After the simulation, we provide a comprehensive report detailing the results. This report includes our findings, interpretations, and noteworthy observations from the simulation process. Our aim is to give you a clear and detailed understanding of the substrate binding process in your specific biological system.
We're here to assist you. If you have any questions, need more information, or would like to discuss a potential project, please don't hesitate to contact us. Our team is always eager to help and share our expertise.
Applications
| Application |
Description |
| Enzyme Engineering |
Enables the prediction of substrate binding pathways using computational simulations. This aids in understanding enzyme-substrate interactions and guides the design of enzymes with optimized binding properties for various biotechnological applications. |
| Drug Discovery |
Facilitates the prediction of substrate binding pathways to target enzymes, aiding in virtual screening of compound libraries and rational drug design. This approach enhances the efficiency of drug discovery by providing insights into potential drug interactions and guiding the selection of lead compounds for further development. |
| Protein-Ligand Docking |
Facilitates protein-ligand docking studies by predicting substrate binding pathways through simulation. This enables the identification of potential binding modes and binding sites on target proteins, aiding in drug discovery and structure-based drug design. |
| Enzyme Kinetics Studies |
Supports enzyme kinetics studies by simulating substrate binding pathways, providing insights into the mechanisms of enzyme-substrate interaction and catalytic activity. This aids in the characterization of enzyme kinetics parameters and the optimization of enzyme performance for industrial applications. |
| Metabolic Pathway Analysis |
Contributes to metabolic pathway analysis by predicting substrate binding pathways for enzymes involved in biochemical reactions. This aids in understanding metabolic fluxes and regulation, guiding metabolic engineering efforts for the production of biofuels, pharmaceuticals, and other valuable compounds. |
FAQs
The following represents a series of frequently asked questions and their respective answers, providing further details about the Predicting Substrate Binding Pathways through Simulation technique:
Q: Could you elaborate on what a substrate means in the context of this technique?
A: Certainly, in the context of this technique, a substrate is essentially a molecule or an ion that has the ability to bind to a predetermined site within a biological system, thereby playing a crucial role in the biological reactions that take place.
Q: Can you comment on the accuracy of the simulations that you conduct?
A: Absolutely, we employ advanced computational models in our work, which allow us to provide predictions that are not just approximations, but are highly accurate and reliable. This high level of accuracy is a testament to the sophistication and precision of our computational models.
Q: Would this technique be useful in the field of drug discovery?
A: Absolutely, the technique of Predicting Substrate Binding Pathways through Simulation proves to be an invaluable tool in the field of drug discovery. It enables us to analyze and predict how potential drug molecules might interact with their target sites in the body.
Q: Could you give some examples of biological systems that can be studied using this technique?
A: The beauty of this technique lies in its high versatility. It can be applied to study any biological system where substrate binding occurs, making it a universally applicable technique in the study of biological interactions.
Q: How long does a typical simulation run for?
A: The duration of a simulation can vary significantly, typically depending on the complexity of the biological system that is being modeled. More complex systems may require longer simulation times to accurately represent the system's behavior.
Q: Can you discuss some of the advantages of using computational modeling?
A: One of the key advantages of computational modeling is that it allows us to predict and understand complex biological processes with a high degree of precision. By using computational models, we eliminate the need for costly and time-consuming laboratory experiments, thereby streamlining the research process.
Q: What kind of information or data is required to run these simulations?
A: In order to run these simulations, we generally need structural data pertaining to the biological system and the substrate. This includes details about the molecular structures of the components involved and their interactions within the system.
Q: Is it feasible to model the simultaneous binding of more than one substrate?
A: Absolutely, our models are designed to handle complex situations, including the simultaneous binding of multiple substrates. This allows us to study more complex biological systems and interactions.
Q: Can this technique be used to study the process of enzyme catalysis?
A: Yes, the technique of Predicting Substrate Binding Pathways through Simulation is an excellent tool for studying enzyme catalysis. It offers insights into the binding and interaction processes of enzymes, thereby aiding in the understanding and prediction of enzyme behavior.

































 Computational design and molecular dynamics simulations suggest the mode of substrate binding in ceramide synthases (Iris D. Zelnik, et al., 2023)
Computational design and molecular dynamics simulations suggest the mode of substrate binding in ceramide synthases (Iris D. Zelnik, et al., 2023)