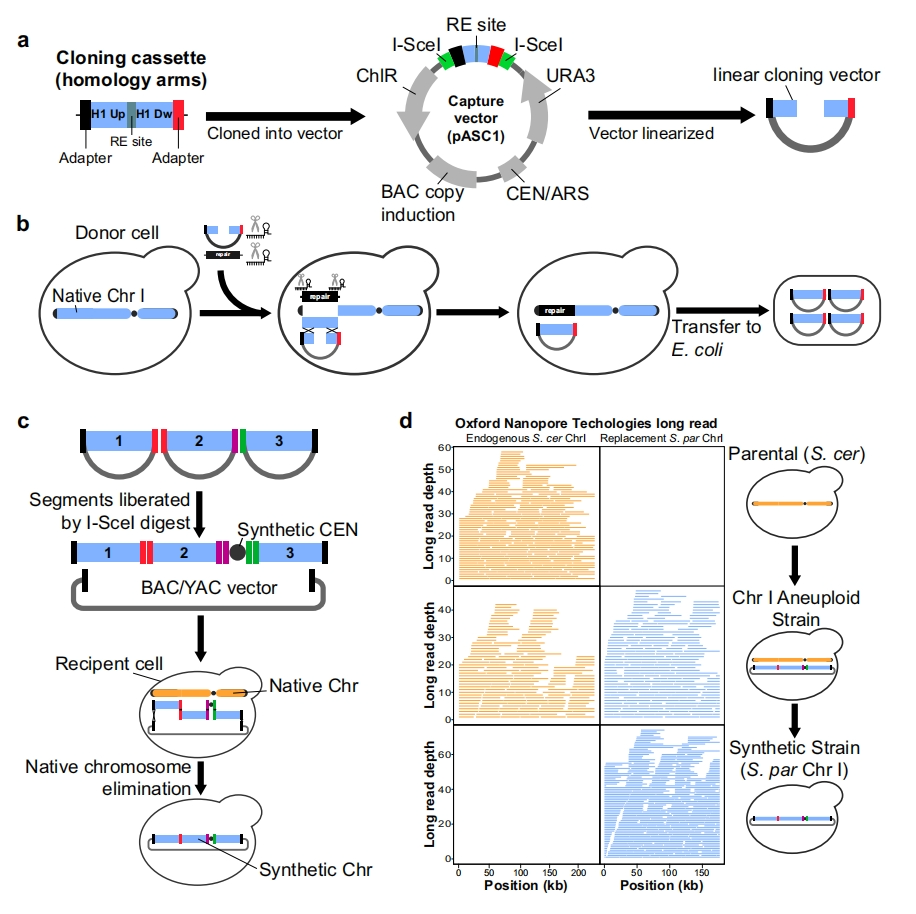
Despite containing only 7.5 artificial chromosomes, highly edited strains still survive and replicate.

Biologists have developed a yeast whose genome contains over 50% of artificially synthesized DNA. Standard brewing yeast (Saccharomyces cerevisiae) stores its genetic blueprint on 16 chromosomes. In the new strain, 6.5 of these chromosomes were edited and synthesized in the laboratory, while the other one was spliced together using edited yeast genetic code fragments.
This article is about the Sc2.0 project, which aims to construct a eukaryotic synthetic genome from scratch. The alliance has been attempting to create a yeast strain with a fully synthetic genome for 15 years.. The article reports an important milestone achieved, where all individual Sc2.0 chromosomes have been assembled. The next step is to consolidate these synthesized chromosomes, which is the focus of this article.
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0011 | High-Fidelity DNA Assembly Cloning Kit | 10 Reactions | Function as a DNA-guided endonuclease. | Online Inquiry |
| GE0012S | High-Fidelity DNA Assembly Master Mix | 10 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012L | High-Fidelity DNA Assembly Master Mix | 50 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
| GE0012X | High-Fidelity DNA Assembly Master Mix | 250 Reactions | High-fidelity construct generation for CRISPR workflows. | Online Inquiry |
A set of papers published in Cell and Cell Genomics describes the team’s achievements, how it manufactured new strains, and other tests it conducted on the yeast genome.
The article describes the integration of multiple synthetic chromosomes using advanced internal replication hybridization and tRNA expression boxes, resulting in a strain with 6.5 synthetic chromosomes. This method can evaluate the profile of 3D chromosome tissue and transcriptional isomers, using Hi-C and long read direct RNA sequencing. The article introduces a new technology called CRISPR directed biallelic URA3 assisted genome scanning, also known as CRISPR D-BUGS. This technique is used to map phenotypic variations caused by specific designer modifications, which are referred to as “bugs”. The article first meticulously mapped a bug in synthetic chromosome II (synII), and then discovered combinatorial interactions related to synIII and synX, revealing an unexpected gene interaction that connects transcriptional regulation, inositol metabolism, and tRNASerCGA abundance. In order to accelerate the integration process, the article used chromosome substitution to integrate the largest chromosome (synIV), thus integrating more than 50% of the Sc2.0 genome in a single strain.
Some viruses and bacteria have been modified into fully synthetic genomes, but they all have simple genetic structures – for example, the bacterium Escherichia coli only has one chromosome. They also have a simple internal structure: for example, bacteria are prokaryotes, which means they are single-celled and do not have a nucleus to accommodate chromosomes. If the Sc2.0 team, consisting of researchers from laboratories in Asia, Europe, North America, and Oceania, can achieve its goals, then their engineered yeast will be the first eukaryotic organism to have a fully synthetic genome. (Eukaryotes are organisms in which cells store genetic material in the nucleus, including humans, plants, and animals.)
The Sc2.0 team hopes to manipulate the synthesized brewing yeast so that one day it can produce drugs and fuel instead of beer. But there are other benefits to this exploration, said Eff Boeke, a synthetic biologist at New York University and the project leader. By adjusting organisms without interfering with their survival, “we have gained a lot of knowledge about the biology of yeast.”.
Nili Ostrov is the Chief Scientific Officer of Cultivarium. Cultivarium is a non-profit company located in Waterton, Massachusetts, specializing in developing tools for bioengineers. He said that Sc2.0 is pushing the limits we can achieve in the field of biotechnology. From a historical perspective, genetic engineers have always focused on modifying individual genes in organisms. Now, biologists can see what happens when they redesign the entire chromosome. Ostrov said, “This allows you to ask some questions that you couldn’t ask before.” She said that in the process, the project is developing methods that can advance biotechnology.
Related Services
Rewrite genome
One of the main goals of Sc2.0 is to eliminate potential sources of instability in the yeast genome. One source is a large amount of repetitive DNA that does not encode anything, but can recombine with each other through natural processes, leading to significant structural changes in the genome. Synthetic biologists wanted to have complete control over their engineered yeast, so this team used computer programs to comb through the genome of brewing yeast, identify highly repetitive regions – and then delete them. Boeke said that these sequences are actually “genomic parasites”.
Another change made by researchers to reduce instability is to remove all DNA fragments encoding transfer RNA from chromosomes and relocate them to fully synthesized “new chromosomes” 2. Transport RNA (tRNA) is crucial for the function of cells – they transport amino acids to a device that uses these molecules to manufacture proteins. But the DNA sequences encoded for them are unstable hotspots. Therefore, transferring them to their own new chromosomes for greater stability is a way for the research team to better control the synthesis of yeast and explore biological limits.
In order to integrate 7.5 synthetic chromosomes into a single cell, the research team created yeast strains, each containing an edited chromosome and 15 other natural versions of chromosomes. Then, they cultivated two strains and selected offspring containing two different edited chromosomes. Then, these strains were cultured and added to another edited chromosome, and so on.
Boeke said that even if chromosomes undergo significant changes, cells with 7.5 chromosomes ultimately survive and can replicate. Although the process of manufacturing cells is time-consuming, what truly slows down is debugging. Researchers must first test whether each yeast cell containing newly synthesized chromosomes has vitality – which means it can survive and function normally – and then solve any problems by adjusting the genetic code. When two or more synthetic chromosomes are located in the same cell, this may lead to new errors that must be repaired, making debugging issues more complex as the process progresses.
Ostrov said that the Sc2.0 project allows scientists to test previously impossible problems, such as, “What happens when you introduce chromosomes that didn’t exist before?”
Boeke said that the team is now trying to replace the remaining natural chromosomes with fully synthesized chromosomes, adding a new chromosome each time, and then debugging an increasingly complex system. “This will require a lot of work to redo.”
Overall, this article provides important updates on the Sc2.0 project and introduces some innovative technologies in the fields of synthetic biology and genomics. These technologies not only help consolidate the synthesis of chromosomes, but also provide powerful tools for future research and understanding of the complexity and regulation of eukaryotic genomes. This article is a great example of the progress in scientific research and how new technologies can drive our understanding of life sciences.