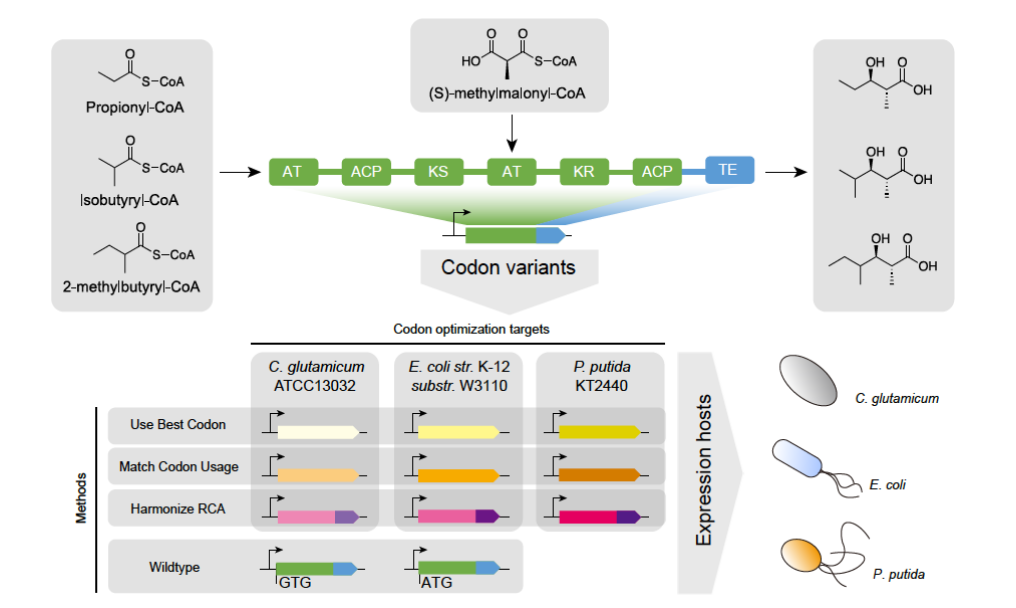
Prime Editing is another technology developed by Professor David R. Liu’s team at Harvard University for precise gene editing, following Base Editing. The development of PE perfectly solves the problem of being able to effectively repair only four gene mutations (i.e. C → T, G → A, A → G, and T → C) in BE, making it repairable for all twelve mutations (the remaining eight being C → A, C → G, G → C, G → T, A → C, A → T, T → A, and T → A). Meanwhile, PE has higher editing efficiency than BE when applied to mutation editing at multiple loci or when bystander editing is not required. Therefore, PE is like a “missile” in gene editing, forming complementary advantages with the “bullet” like BE.
| Catalog Number | Product Name | Product Size | Applications | Price |
|---|---|---|---|---|
| GE0013 | Genomic Cleavage Detection Kit | 25 Reactions | Single-tube, isothermal generation of the Cas9-sgRNA constructs. | Online Inquiry |
| GE0014 | Mutation Detection Kit | 25 Reactions | Determination of the targeting efficiency of genome editing. | Online Inquiry |
| GE0015S | T7 Endonuclease I | 250 Units | Function as a DNA endonuclease. | Online Inquiry |
| GE0015L | T7 Endonuclease I | 1,250 Units | Function as a DNA endonuclease. | Online Inquiry |
| GE0016 | Tth Argonaute | 50 pmol | Single-tube, isothermal generation of the Cas9-sgRNA constructs. | Online Inquiry |
The Infrastructure of PE and PE1
Prime Editing relies on the implementation of the Prime Editor. The main components of Prime Editor (PE) are RNA programmable scratch enzyme (nCas9) fused reverse transcriptase (RT) and primary editing guide RNA (pegRNA). PegRNA is composed of a spacer with a specified target site, an sgRNA scaffold, and a primer binding site (PBS) of 3 ‘that needs to be edited.

Schematic diagram of PE structure
PE1: PE1 is the prototype of the aforementioned PE structure. So how does PE1 perform its gene editing function? There are roughly the following steps in the process:
Binding: The pegRNA complex binds to the target site on the target DNA and cleaves the PAM containing strand.
Reverse transcription: The obtained cut DNA strand base pairs are transferred to PBS in pegRNA, and reverse transcription of the RT template is initiated directly into the target DNA site.
Repair: Obtain the newly synthesized 3 ‘strand of edited DNA through the cellular DNA repair pathway, thereby completing the required editing at the target site.

Schematic diagram of the working principle of PE1
PE’s”Evolution”
PE2: Based on PE1, the experimenters developed PE2. Compared to PE1, the improvement of PE2 lies in the use of engineered RT or M-MLV RT mutants to replace the original RT. The advantages of the M-MLV RT variant lie in its better thermal stability, synthesis, and DNA: RNA affinity. Through experimental characterization, it was found that the editing efficiency of PE2 is much higher than that of PE1, and it is compatible with shorter PBS sequences.

Comparison of editing efficiency between PE1 and PE2
PE3: As nCas9 can only cleave one homologous chain, in PE1 and PE2, there is one edited chain and one unedited chain. If the unedited chain can be cut off to form two edited chains, will editing efficiency be improved? Following this train of thought, PE3 was born out of nowhere.
In PE3, the experiment used Cas9 H840A cleavage enzyme already present in PE2 and simple sgRNA to cut the unedited strand, and used the edited strand as a template to direct DNA repair to that strand. A series of experiments have shown that PE3 improves editing efficiency by about three times compared to PE2.

PE3 design concept diagram
PE3b: Scratches should be made on unedited chains only after they have been edited, and the presence of concurrent scratches should be minimized to minimize chain breakage and depression formation. Therefore, based on PE3, PE3b was experimentally designed. PE3b reduces breaks and concavities by designing sgRNAs that match the spacer with the edited strand, but do not match the original allele. Compared with PE3, the average insertion rate of PE3b decreased by 13 times, but at the same time, there was no significant decrease in editing efficiency.

Comparison of editing efficiency between PE2, PE3, and PE3b
In PE2, the structure of pegRNA, PBS length, and RT template length can to some extent affect editing efficiency. The development of PE3 enables long-distance RT templates to generate high editing rates without being substantially limited by the availability of nearby PAM sequences. Therefore, the developed PE3 has multifunctionality, precision, specificity, and flexibility.
Comparison of PE with BE and HDR (homologous repair technology)
The experiment compared PE and BE editing techniques and found that when there is a single target nucleotide in the BE window or when bystander editing requires it, BE is usually more effective and produces less embedding than PE. But when there are multiple cytosines or adenines that do not require bystander editing, or when PAM for base editing targeting nucleotides is not available, PE provides a more substantial advantage.
Therefore, PE and BE can provide complementary advantages.

Comparison of Editing Efficiency between PE and BE
The experiment also compared HDR, a gene editing technique, and found that PE has lower levels of byproduct production, lower off target rates, and higher editing efficiency compared to HDR.
Therefore, PE has significant advantages over HDR.

Comparison of editing efficiency between PE and HDR
Conclusion
Developed three types of Prime Editing editors, namely PE1, PE2, and PE3.
Prime Editing provides many possible positions for pegRNA induced incisions, the second position for sgRNA induced incisions, PBS length, RT template length, and the first edited strand. Therefore, it has strong flexibility and is in sharp contrast to the more limited options typically available in other precise editing methods. It allows for optimization of editing efficiency, product purity, DNA specificity, and other parameters to adapt to a given application.
PE and Its Applications and the Future
Like BE, the development of PE is mainly aimed at applying it to the treatment of single gene mutation gene diseases. The use of PE in the treatment of sickle cell disease (etiology: A • T-to-T • A in bromodiphenyl) and Tay Sachs disease (etiology: 4 bp insertion in HEXA) has achieved good results in the article.
Of course, the iteration of PE technology does not stop at PE3. Last year, David The Liu team has developed PE4 (PE2+MLH1dn) and PE5 (PE3+MLH1dn) systems to address the issue of increased frequency of insertion or deletion (indel) of unwanted bases when removing unedited chains. Among them, MLH1dn is a dominant negative MMR (DNA mismatch repair) protein, which inhibits MMR through its transient expression, thereby reducing indel production and improving editing efficiency.
This year, David The Liu team further optimized the PE system using engineered pegRNA (epegRNA) containing structured RNA pseudojunctions, targeting PBS in the 3 ‘extension of PEgRNA that is easily degraded by exogenous enzymes. This has once again improved the efficiency of editing.
In short, gene editing technology is still constantly evolving, and PE technology will also be continuously improved and optimized over time. Gene editing technology, the future is promising!
The original text is from Andrew V. Anzalone, 1,2,3 Peyton B. Randolph, 1,2,3 Jessie R. Davis, 1,2,3 Alexander A. Sousa, 1,2,3 Luke W. Koblan, 1,2,3 Jonathan M. Levy, 1,2,3 Peter J. Chen, 1,2,3 Christopher Wilson, 1,2,3 Gregory A. Newby, 1,2,3 Aditya Ragram, 1,2,3 and David R. Liu1,2,3, *, Search and replace gene editing without double stra D breaks or donor DNA, [J], Nature 2019 Dec; 576 (7785): 149-157