The protein engineering and drug discovery domains have undergone transformative changes due to mRNA display and ribosome display technologies which allow scientists to study protein functionality and evolve enzymes while identifying novel therapeutic agents. These techniques allow for the examination of extensive protein and peptide libraries although their operational principles and practical uses show significant differences. Researchers seeking a detailed comparison between mRNA display and ribosome display will find this article examines the essential principles and workflows while highlighting the strengths of each technique.
1. Introduction
Modern biotechnology techniques depend fundamentally on connecting genetic information to the proteins or peptides they encode. The technologies mRNA display and ribosome display produce extensive protein libraries and enable selection of proteins based on key characteristics like binding affinity or enzymatic performance. Although both technologies share comparable objectives they employ different technical methods and demonstrate varying levels of effectiveness for specific applications.
Navigating between these technologies requires researchers to comprehend their unique features. The decision between mRNA display and ribosome display affects both the efficiency and cost of experiments while determining the ultimate success of antibody optimization, enzyme engineering, or ligand development projects. The article analyzes the fundamental distinctions between technologies alongside their applications and important factors to assist your decision-making.

Comparison of two alternative strategies for coupling phenotype and genotype in vitro (Lipovsek, et al. 2004)
2. Understanding mRNA Display
Core Principle
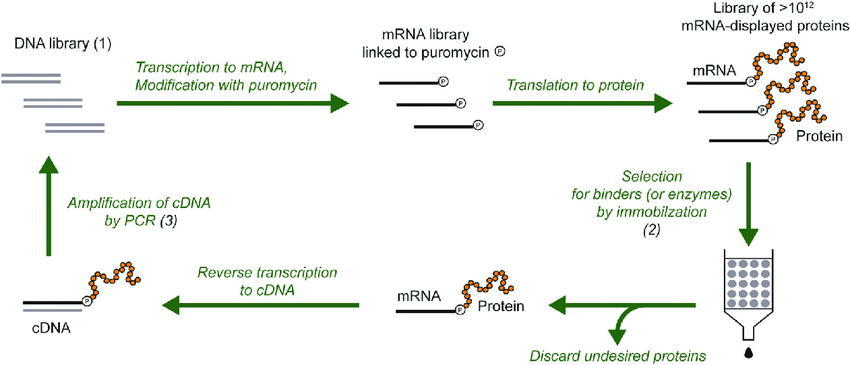
The mRNA display technique enables the creation of stable complexes between mRNA molecules and the peptides or proteins they encode during in vitro selection processes. A non-covalent interaction between mRNA and the peptide establishes this linkage while being stabilized by the presence of a puromycin molecule. The antibiotic puromycin acts as a substitute for the 3’ end of tRNA and binds covalently to the peptide chain as it develops during the translation process. The resulting mRNA-peptide conjugate remains stable which permits direct connection between the genotype represented by the mRNA and the resulting phenotype expressed as peptide or protein. Explore mRNA Display Screening Services provided by Biosynsis.
Workflow
1.Library Construction: The mRNA is produced through transcription from a DNA library that contains specific peptides or proteins.
2.In Vitro Translation: Scientists use cell-free translation systems including rabbit reticulocyte lysate or Escherichia coli extract to translate mRNA. The translation process ends prematurely when puromycin joins the mRNA to the nascent peptide through covalent bonding.
3.Complex Stabilization: The mRNA-peptide complexes are stabilized and purified.
4.Selection: The process involves screening the complexes against a target such as a receptor, enzyme, or antibody to select those which demonstrate high binding affinity.
5.Recovery and Amplification: The selected mRNA undergoes reverse transcription to produce cDNA which researchers amplify by PCR to create a new library for further selection rounds.
Strengths
- Large Library Sizes:The mRNA display method supports libraries containing up to 10¹⁵ unique sequences which results in outstanding sequence diversity.
- Eukaryotic Compatibility:Eukaryotic cell-free systems such as rabbit reticulocyte lysate enable post-translational modifications which makes them appropriate for eukaryotic proteins.
- Flexible Screening Conditions:Because the mRNA-peptide complex maintains non-covalent interactions it enables screening across diverse conditions ranging to denaturing environments.
3. Understanding Ribosome Display
Core Principle
During in vitro translation ribosome display requires the creation of a stable ternary complex that consists of mRNA and a ribosome paired with the growing peptide chain. Translation arrest at precise points through methods such as termination codon omission or application of translation inhibitors like cycloheximide keeps the ribosome attached to the mRNA along with the incomplete peptide. The structural complex forms a direct connection between the genetic sequence (mRNA) and its resulting structure (peptide), which allows for selection to occur. For comprehensive ribosome display service, visit Ribosome Display Service.
Workflow
1.Library Construction: The transcription process converts DNA libraries that lack stop codons into mRNA sequences.
2.In Vitro Translation: Cell-free translation of mRNA generates a collection of ribosome-mRNA-peptide complexes. To preserve the ternary complex ribosome-mRNA-peptide structure scientists stop translation midway.
3.Selection: During incubation with a target molecule the complexes containing high-affinity peptides remain bound.
4.Recovery and Amplification: After extracting the selected mRNA from ribosome complexes scientists convert it to cDNA through reverse-transcription before using PCR to amplify it for future selection processes.
Strengths
- Simplicity and Cost-Effectiveness: Ribosome display removes dependence on puromycin and linker molecules resulting in reduced complexity and lower costs for the workflow.
- Efficient Screening: The stable nature of the ribosome-mRNA-peptide complex facilitates efficient selection of high-affinity binders.
- Large Library Handling: Ribosome display matches mRNA display in its ability to handle up to 10¹³–10¹⁴ sequences which provides sufficient diversity for most applications.
4. mRNA Display vs. Ribosome Display: A Detailed Comparison
This table summarizes the technical mechanisms, capabilities, and limitations of both technologies to demonstrate their key differences:
| Feature | mRNA Display | Ribosome Display |
| Linkage Mechanism | Non-covalent mRNA-peptide complex via puromycin | Physical ribosome-mRNA-peptide ternary complex |
| Translation System | Eukaryotic or prokaryotic cell-free lysates | Prokaryotic cell-free lysates (e.g., E. coli) |
| Library Size | Up to 10¹⁵ sequences | Up to 10¹³–10¹⁴ sequences |
| Protein Types | Suitable for eukaryotic proteins with post-translational modifications | Primarily for peptides and prokaryotic proteins |
| Screening Conditions | Flexible, including denaturing environments | Limited to native or near-native conditions |
| Complex Stability | Moderate stability; sensitive to harsh conditions | Highly stable; withstands stringent washing |
| Cost and Complexity | Higher cost due to puromycin and eukaryotic systems | Lower cost; simpler workflow |
| Throughput | High, suitable for large-scale screens | High, but slightly lower than mRNA display |
Key Takeaways from the Comparison
- Technical Mechanisms: The mRNA display technique utilizes puromycin as a chemical linker between mRNA and peptide whereas ribosome display depends on physical ribosome complexes.
- Protein Diversity: Eukaryotic proteins needing post-translational modifications perform better with mRNA display but ribosome display suits peptides and basic prokaryotic proteins more effectively.
- Screening Flexibility: The mRNA display technique functions effectively under harsh screening conditions while ribosome display performs best in natural environmental settings.
- Cost and Workflow:The cost-effective simplicity of ribosome display makes it the preferred option for laboratories that need to manage their budgets effectively.
5. mRNA Display vs. Phage Display: A Quick Contrast
In many cases, mRNA or ribosome display may be preferable for high-throughput screens or eukaryotic protein engineering.
This article examines mRNA display and ribosome display yet a short comparison of these technologies with phage display helps to contextualize their advantages. Phage display connects proteins and bacteriophage genomes by attaching them to phage coat proteins. Here’s how mRNA and ribosome display differ:
- Speed:The advantage of mRNA and ribosome display lies in their expedited processes which eliminate the necessity for bacterial infection and subsequent amplification steps.
- Library Size: The technology of mRNA display produces significantly larger libraries than the traditional phage display approach as evidenced by a difference from 10¹⁵ to a range of 10¹¹–10¹².
- Eukaryotic Proteins: Eukaryotic proteins present challenges for phage display because bacterial systems cannot properly fold them while mRNA display successfully manages them through eukaryotic lysates.
High-throughput screening and eukaryotic protein engineering often benefit from the use of mRNA and ribosome display methods.
6. Applications of mRNA and Ribosome Display
Biotechnology and drug discovery fields have widely adopted both technologies for various applications. The following segment illustrates practical applications of both technologies.
mRNA Display
- Antibody Discovery:Scientists use mRNA display to create extensive antibody libraries for screening and isolating high-affinity antibodies that target difficult antigens. Explore mRNA Display – based Antibody Discovery to see how this process is carried out in detail.
- Enzyme Evolution:Through the screening of enzyme variants scientists manage to develop catalysts which demonstrate better performance in terms of activity as well as stability and specificity. Learn more about mRNA Display – based Enzyme Directed Evolution for in – depth knowledge.
- Ligand Binding Studies:The mRNA display technique assists scientists in discovering binding peptides and small proteins that attach to particular receptors or enzymes which supports drug development efforts.

Antibody-selection strategy in mRNA display(Lipovsek, et al. 2004)
Ribosome Display
- Peptide Library Screening:Scientists frequently utilize ribosome display to screen peptide libraries for potential ligands and either inhibitors or agonists. If you’re interested in peptide discovery, check out mRNA Display – Based Peptide Discovery at CD Biosynsis.
- Affinity Maturation:The binding affinity of antibodies and peptides gets improved through iterative selection of variants by researchers. Enhance antibody affinity via mRNA Display – Based Antibody Affinity Maturation.
- Directed Evolution:The ribosome display technique speeds up protein evolution toward specific properties including improved solubility and thermal stability. Uncover novel proteins with mRNA Display – Based Protein Discovery from CD Biosynsis.
7. Choosing Between mRNA Display and Ribosome Display
Your research objectives combined with the protein type and available resources determine the technology selection process. Here’s a decision-making guide:
When to Choose mRNA Display
- Eukaryotic Proteins:The eukaryotic translation systems of mRNA display become necessary when your target protein requires post-translational modifications like glycosylation. Screen proteins with mRNA Display – Based Protein Screening.
- Extreme Screening Conditions:Should your experiments require protein screening under conditions that denature proteins or create non-native environments? mRNA display complexes possess flexibility that allows them to endure more extreme conditions. Explore the development of unnatural proteins with mRNA Display – Based Unnatural Protein Development.
- Very Large Libraries: When libraries surpass 10¹⁴ sequences mRNA display becomes the preferred choice due to its ability to handle 10¹⁵ sequences and deliver unmatched diversity.Create self – assembling protein microarrays through mRNA Display – Based Self – Assembling Protein Microarray Development.
When to Choose Ribosome Display
- Cost and Simplicity:Labs operating under budget restrictions benefit from ribosome display because of its cost-effective and streamlined workflow.
- Peptide or Prokaryotic Protein Focus:Ribosome display proves efficient and effective for projects involving well-folding peptides and proteins in prokaryotic systems.
- Stable Complexes: The ribosome-mRNA-peptide complex demonstrates high stability which enables rigorous washing throughout the selection process.
Other Considerations
- Downstream Applications:The compatibility of mRNA display with eukaryotic systems offers benefits when expressing selected proteins in eukaryotic cells.
- Throughput: Although both technologies can handle high throughput operations, mRNA display comes out ahead for ultra-large screening purposes.
8. Conclusion
The techniques of mRNA display and ribosome display serve as effective methods for advancing protein engineering and drug discovery because they provide distinct capabilities. The mRNA display technology stands out for its capability to process eukaryotic proteins and manage large libraries under flexible screening conditions whereas ribosome display provides benefits through its simple operation, cost-efficiency, and stable performance. Your research goals whether they pertain to antibody development enzyme evolution or peptide screening will guide you to choose the most appropriate technology. Drive enzyme – directed evolution with mRNA Display – Based Enzyme – Directed Evolution.
Selecting between mRNA display and ribosome display depends on evaluating technical needs alongside project challenges and available resources. The ongoing development of these technologies keeps them essential for advancing protein science and therapeutic discovery.
To gain additional insight you should engage with specialists and examine case studies about technology implementation in comparable research situations. Applying the correct methodology enables you to maximize mRNA or ribosome display capabilities in your scientific endeavors.
Reference
Lipovsek, Dasa, and Andreas Plückthun. “In-vitro protein evolution by ribosome display and mRNA display.” Journal of immunological methods 290.1-2 (2004): 51-67.