Phage-Assisted Continuous Evolution (PACE) Technology
- Home
- EnzymoGenius™ Technology
- Phage-Assisted Continuous Evolution (PACE) Technology
































Phage-Assisted Continuous Evolution (PACE) technology is a revolutionary method of directed protein evolution. The technology was developed by Liu Ruqian's team at Harvard University in 2011 and aims to generate protein or RNA molecules with new functions through a rapid and continuous evolutionary process. Compared with traditional laboratory evolution methods, PACE can achieve hundreds of rounds of evolution in a short time, significantly improving research efficiency.
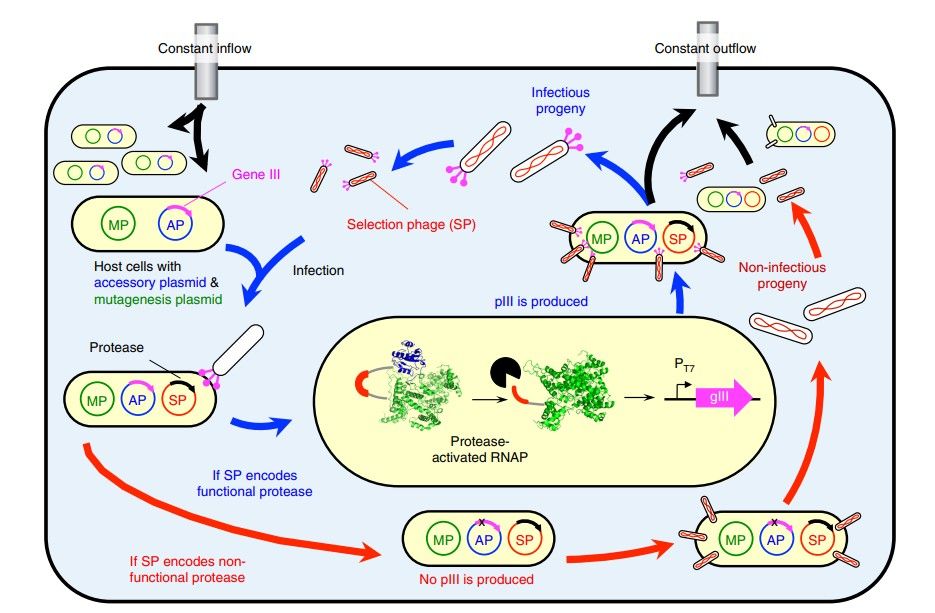 Overview of protease PACE(MS Packer, et al., 2017)
Overview of protease PACE(MS Packer, et al., 2017)
The core of PACE technology is to use phage (a virus that infects bacteria) as a vector to drive the directed evolution of target genes through evolving phage. Specifically, the system relies on a continuously flowing "pond" environment that contains M13 bacteriophages (select phage SP) that rely on protein III (pIII) in host cells to grow. The production of Protein III is directly related to the activity of the target gene. Only when the target gene exhibits the desired function can the phage survive in the pond and continue to replicate.
As part of PACE, functionally better variants of proteins are continuously weeded out by random mutations and selection pressure. This method not only accelerates protein evolution but also eliminates the need for humans and can streamline the process of experimental design. Additionally, PACE technology is extremely versatile and can be leveraged to design a wide array of biological molecules, such as enzymes, antibodies or gene editing platforms.
As a CRO service outsourcing company, we provide advanced PACE technical support to help customers achieve breakthroughs in the biotechnology field. Whether it's accelerating new drug development, optimizing biocatalysts, or developing new gene editing tools, our PACE technology can provide strong support for your project. By choosing our services, you will experience unprecedented research efficiency and innovation potential.
Fast evolution speed
PACE technology can execute up to 60 rounds of evolution per day, dramatically outperforming lab-based evolution.
No manual intervention required
The entire experiment doesn't need frequent human intervention, thereby saving money and time.
Wide application range
PACE technology has been used for the evolutionary modification of RNA polymerase, TALEN, Cas9 and other enzymes.
High-throughput screening
Around 200 generations of directed evolution can be performed per week, which allows for large-scale screening.
Low cost and easy to operate
The system is easy to set up and inexpensive enough to be useful for both hobbyist biologists and small labs.
Efficient mutation generation
By using phage-guided continuous evolution, many mutants with desired traits can be generated quickly.
Reduce human error
Since the evolution process is largely automated, human-generated mistakes and incongruencies are minimized.
Highly flexible
Appropriate for directed evolution of many proteins and nucleic acids, including those requiring complex folding or post-translational modifications.
Phage-assisted continuous evolution (PACE) is an effective strategy for targeted evolution of biological molecules, a method in many fields. The technology achieves quick and efficient evolution of the target protein by coupling the target protein activity with the expression of pIII, one of the phage's most important proteins. Specific application areas include:
PACE technology is capable of rapidly developing therapeutic proteins and enhance the function and stability of proteins through targeted evolution.
Gene editing tools such as Cas9 variants are being optimised for increased PAM compatibility and DNA specificity with the help of PACE technology.
PACE can quickly engineer targeted enzymes — for example, RNA polymerases, proteases etc — to improve their catalytic efficiency and specificity.
Screening of chemical synthesis enzymes
PACE technology enables enzymes with high chemical synthesis efficiency to be screened which will boost chemical synthesis efficiency.
Agricultural biotechnology
Using PACE technology, researchers can create novel insecticidal proteins to overcome resistance.
Biosensor design
PACE is capable of designing sensitive, high-specificity biosensors to identify certain molecules or environmental changes.
Here are answers to frequently asked questions (FAQs) about Phage-Assisted Continuous Evolution (PACE) Technology
Q: What is PACE technology?
A: PACE (Phage-Assisted Continuous Evolution) is a technology that uses bacteriophages as carriers to rapidly evolve proteins through a continuous process of mutation and selection. The technology does not require DNA library design, cell transformation, gene extraction or cloning in the traditional sense, so it performs multiple rounds almost "autonomous", allowing up to 38 generations of mutations and selections per day.
Q: How does PACE technology work?
A: In the PACE system, the M13 phage (select phage SP) relies on the production of protein III (pIII) for growth. These bacteriophages were modified so that pIII production was correlated with protein activity. When the protein encoded by SP is active, pIII is produced from the helper plasmid (AP), allowing the phage displaying the protein to replicate and survive in a continuously flowing environment.
Q: How is PACE technology different from traditional directed evolution methods?
A: Compared to traditional directed evolution methods, PACE technology does not require human intervention and can complete up to 30 evolutionary cycles in one day, while traditional methods usually take several days. In addition, PACE technology achieves uninterrupted evolution through a continuous flow system, allowing about 200 generations of directed evolution per week.
Q: What are the advantages of PACE technology?
A: The main advantages of PACE technology include reducing research workload, eliminating the need for complex DNA manipulation steps, and being able to carry out a large number of mutations and selections in a short period of time, thereby accelerating the evolution of proteins.
Q: How is PACE technology different from other phage-assisted evolution technologies?
A: The main difference between PACE technology and PANCE (discontinuous phage-assisted evolution) is that PANCE requires continuous dilution rather than continuous flow, while PANCE also supports multiple experiments, but at a slower speed. In addition, PRANCE (Near Continuous Phage-Assisted Evolution) is an automated system that can perform phage-assisted evolution at high throughput and has the ability to monitor biological activity in real time.
Q: What is the application scope of PACE technology?
A: PACE technology is widely used in protein engineering, drug development, enzyme engineering and other fields. For example, it can be used to develop new cancer drugs, biofuel production, and synthesize complex compounds. In addition, PACE is also used to modify RNA polymerases to recognize different promoters.
Q: What is the operating process of PACE technology?
A: In the PACE system, the M13 phage infects Escherichia coli cells, and the gene pIII required by the phage is removed from the phage genome and encoded on a plasmid in Escherichia coli. When a phage infects Escherichia coli, if the phage carries a functional protein capable of producing enough pIII, the phage will continue to replicate and infect other host cells.
Q: What limitations should I pay attention to when using PACE technology?
A: There are several limitations to note when using PACE technology: (i) the production of pIII must be associated with a protein target (POI);(ii) the gene encoding the POI must be less than 5 kb in length;(iii) proteins that require PTMs or disulfide bond folding are not suitable for use with this technology because protein production occurs in the cytoplasm of Escherichia coli.
Q: How to evaluate the effectiveness of PACE technology?
A: In the PACE experiment, researchers can assess evolutionary effects by analyzing changes in gene sequence that occur during the mutation process. For example, by testing phage genomic sequences of different generations, it is possible to understand which mutations have a positive impact on the function of the target protein.
Please take a moment to fill out the form.
CD Biosynsis
Copyright © 2025 CD Biosynsis. All rights reserved.
